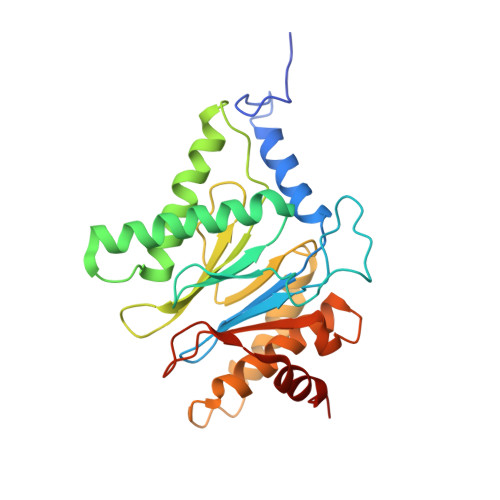
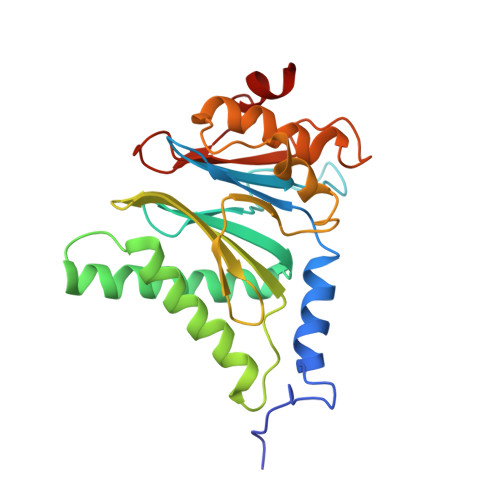
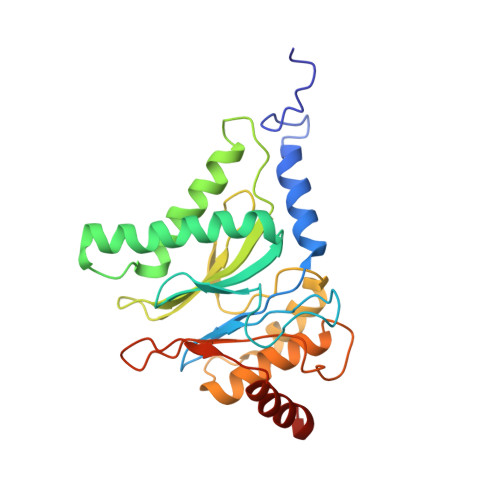
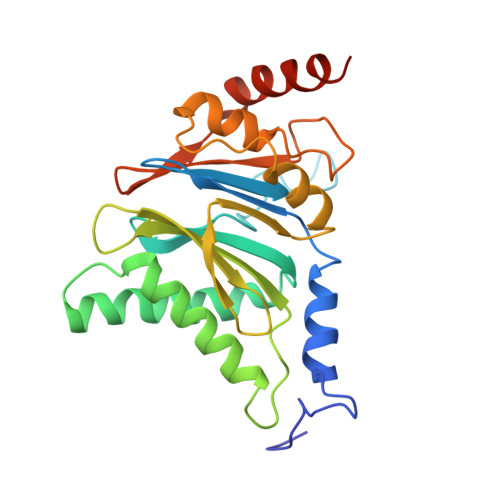
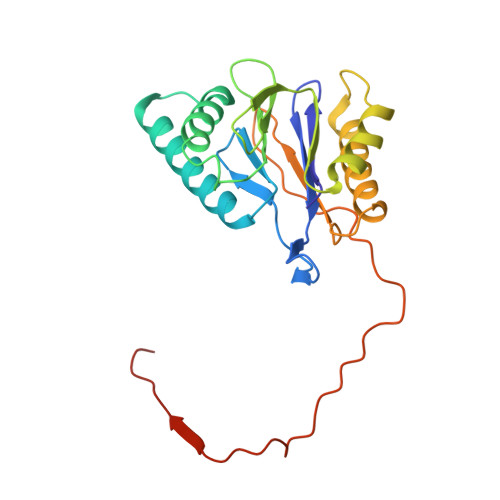
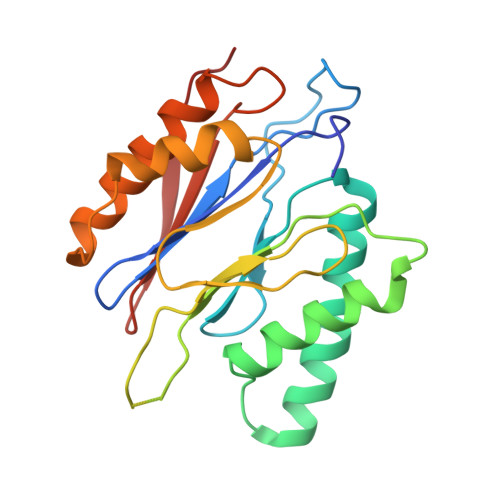
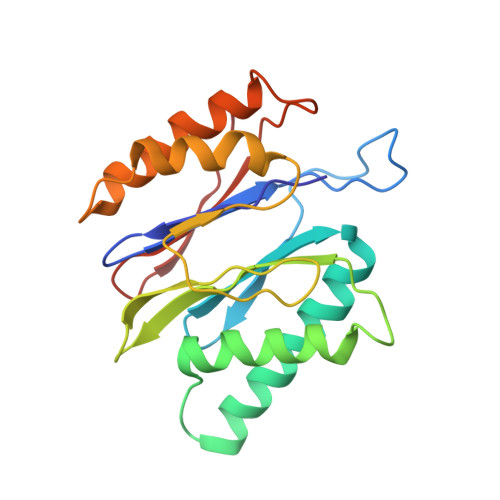
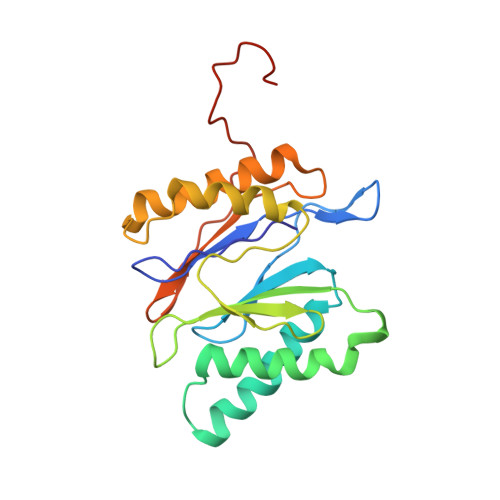
The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Xie, S.C., Metcalfe, R.D., Hanssen, E., Yang, T., Gillett, D.L., Leis, A.P., Morton, C.J., Kuiper, M.J., Parker, M.W., Spillman, N.J., Wong, W., Tsu, C., Dick, L.R., Griffin, M.D.W., Tilley, L.(2019) Nat Microbiol 4: 1990-2000
- PubMed: 31384003
- DOI: https://doi.org/10.1038/s41564-019-0524-4
- Primary Citation of Related Structures:
6DFK, 6MUV, 6MUW, 6MUX - PubMed Abstract:
The activity of the proteasome 20S catalytic core is regulated by protein complexes that bind to one or both ends. The PA28 regulator stimulates 20S proteasome peptidase activity in vitro, but its role in vivo remains unclear. Here, we show that genetic deletion of the PA28 regulator from Plasmodium falciparum (Pf) renders malaria parasites more sensitive to the antimalarial drug dihydroartemisinin, indicating that PA28 may play a role in protection against proteotoxic stress. The crystal structure of PfPA28 reveals a bell-shaped molecule with an inner pore that has a strong segregation of charges. Small-angle X-ray scattering shows that disordered loops, which are not resolved in the crystal structure, extend from the PfPA28 heptamer and surround the pore. Using single particle cryo-electron microscopy, we solved the structure of Pf20S in complex with one and two regulatory PfPA28 caps at resolutions of 3.9 and 3.8 Å, respectively. PfPA28 binds Pf20S asymmetrically, strongly engaging subunits on only one side of the core. PfPA28 undergoes rigid body motions relative to Pf20S. Molecular dynamics simulations support conformational flexibility and a leaky interface. We propose lateral transfer of short peptides through the dynamic interface as a mechanism facilitating the release of proteasome degradation products.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, Victoria, Australia.