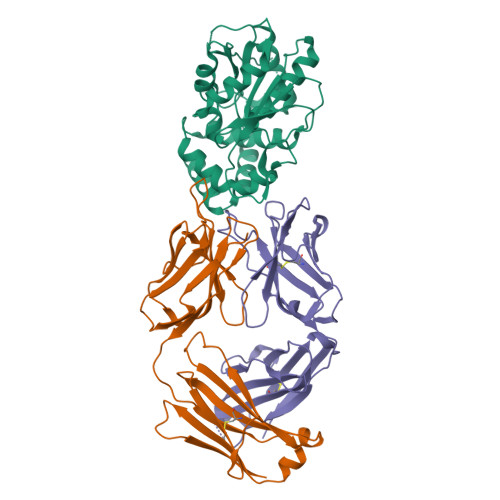
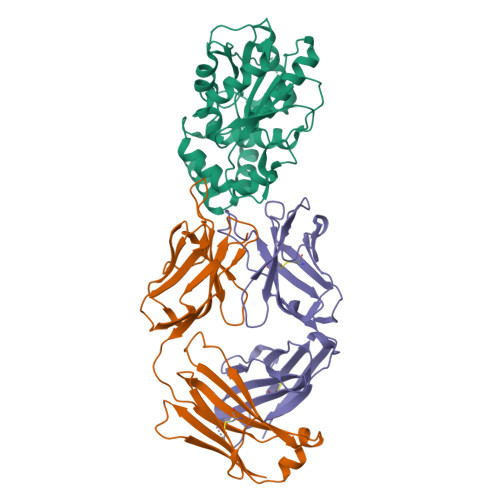
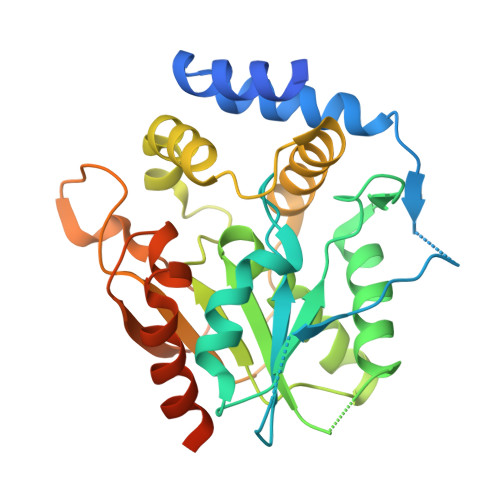
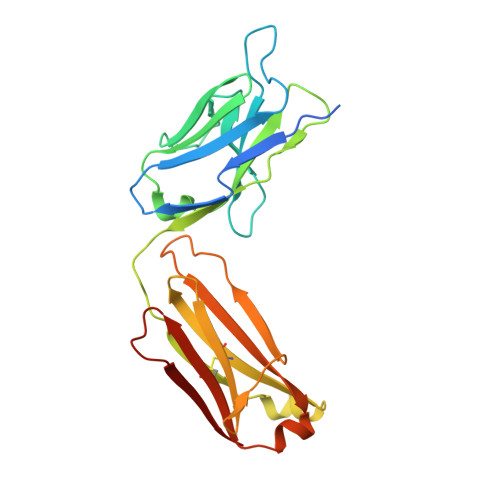
Discovery and Characterization of a Thioesterase-Specific Monoclonal Antibody That Recognizes the 6-Deoxyerythronolide B Synthase.
Li, X., Sevillano, N., La Greca, F., Hsu, J., Mathews, I.I., Matsui, T., Craik, C.S., Khosla, C.(2018) Biochemistry 57: 6201-6208
- PubMed: 30289692
- DOI: https://doi.org/10.1021/acs.biochem.8b00886
- Primary Citation of Related Structures:
6MLK - PubMed Abstract:
Assembly line polyketide synthases (PKSs) are large multimodular enzymes responsible for the biosynthesis of diverse antibiotics in bacteria. Structural and mechanistic analysis of these megasynthases can benefit from the discovery of reagents that recognize individual domains or linkers in a site-specific manner. Monoclonal antibodies not only have proven themselves as premier tools in analogous applications but also have the added benefit of constraining the conformational flexibility of their targets in unpredictable but often useful ways. Here we have exploited a library based on the naïve human antibody repertoire to discover a F ab (3A6) that recognizes the terminal thioesterase (TE) domain of the 6-deoxyerythronolide B synthase with high specificity. Biochemical assays were used to verify that 3A6 binding does not inhibit enzyme turnover. The co-crystal structure of the TE-3A6 complex was determined at 2.45 Å resolution, resulting in atomic characterization of this protein-protein recognition mechanism. F ab binding had minimal effects on the structural integrity of the TE. In turn, these insights were used to interrogate via small-angle X-ray scattering the solution-phase conformation of 3A6 complexed to a catalytically competent PKS module and bimodule. Altogether, we have developed a high-affinity monoclonal antibody tool that recognizes the TE domain of the 6-deoxyerythronolide B synthase while maintaining its native function.
Organizational Affiliation:
Department of Pharmaceutical Chemistry , University of California San Francisco , San Francisco , California 94158 , United States.