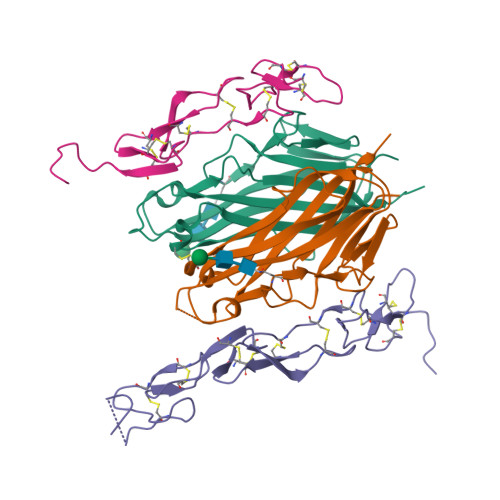
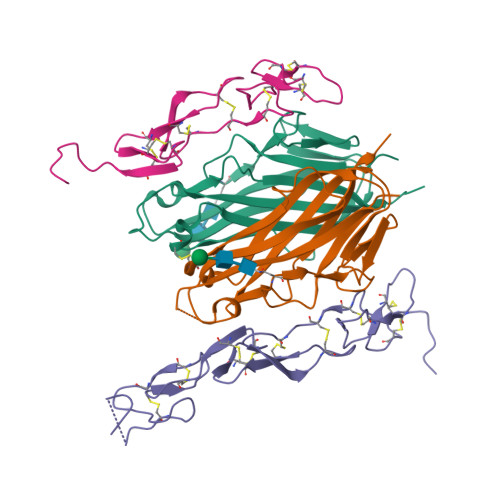
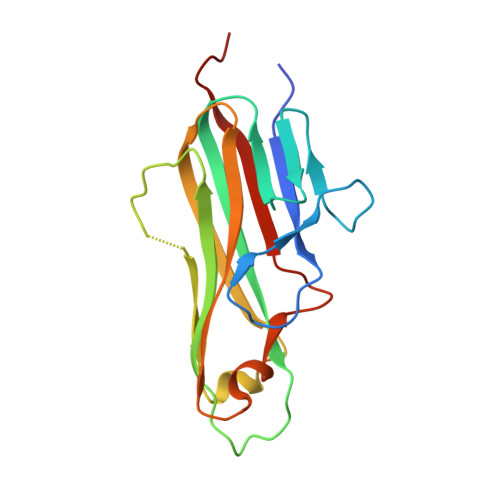
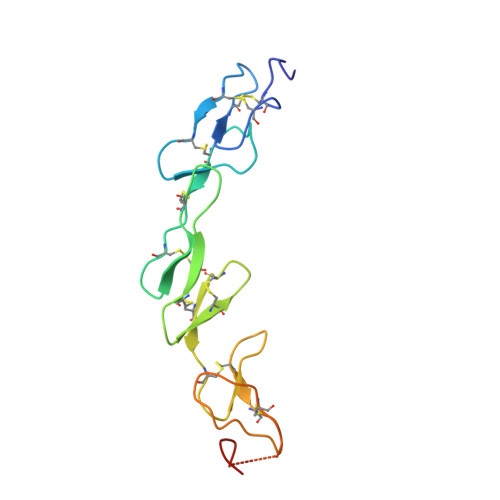
Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
Bitra, A., Doukov, T., Destito, G., Croft, M., Zajonc, D.M.(2019) J Biological Chem 294: 1831-1845
- PubMed: 30545939
- DOI: https://doi.org/10.1074/jbc.RA118.006297
- Primary Citation of Related Structures:
6MKB, 6MKZ - PubMed Abstract:
The interaction between the receptor 4-1BB and its ligand 4-1BBL provides co-stimulatory signals for T-cell activation and proliferation. However, differences in the mouse and human molecules might result in differential engagement of this pathway. Here, we report the crystal structure of mouse 4-1BBL and of the mouse 4-1BB/4-1BBL complex, which together provided insights into the molecular mechanism by which m4-1BBL and its cognate receptor recognize each other. Unlike all human or mouse tumor necrosis factor ligands that form noncovalent and mostly trimeric assemblies, the m4-1BBL structure formed a disulfide-linked dimeric assembly. The structure disclosed that certain differences in the amino acid composition along the intramolecular interface, together with two specific residues (Cys-246 and Ser-256) present exclusively in m4-1BBL, are responsible for this unique dimerization. Unexpectedly, upon m4-1BB binding, m4-1BBL undergoes structural changes within each protomer; moreover, the individual m4-1BBL protomers rotate relative to each other, yielding a dimerization interface with more inter-subunit interactions. We also observed that in the m4-1BB/4-1BBL complex, each receptor monomer binds exclusively to a single ligand subunit with contributions of cysteine-rich domain 1 (CRD1), CRD2, and CRD3. Furthermore, structure-guided mutagenesis of the binding interface revealed that novel binding interactions with the GH loop, rather than the DE loop, are energetically critical and define the m4-1BB receptor selectivity for m4-1BBL. A comparison with the human 4-1BB/4-1BBL complex highlighted several differences between the ligand- and receptor-binding interfaces, providing an explanation for the absence of inter-species cross-reactivity between human and mouse 4-1BB and 4-1BBL molecules.
Organizational Affiliation:
From the Division of Immune Regulation, La Jolla Institute for Immunology (LJI), La Jolla, California 92037.