Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
Da, C., Zhang, D., Stashko, M., Vasileiadi, E., Parker, R.E., Minson, K.A., Huey, M.G., Huelse, J.M., Hunter, D., Gilbert, T.S.K., Norris-Drouin, J., Miley, M., Herring, L.E., Graves, L.M., DeRyckere, D., Earp, H.S., Graham, D.K., Frye, S.V., Wang, X., Kireev, D.(2019) J Am Chem Soc 141: 15700-15709
- PubMed: 31497954
- DOI: https://doi.org/10.1021/jacs.9b08660
- Primary Citation of Related Structures:
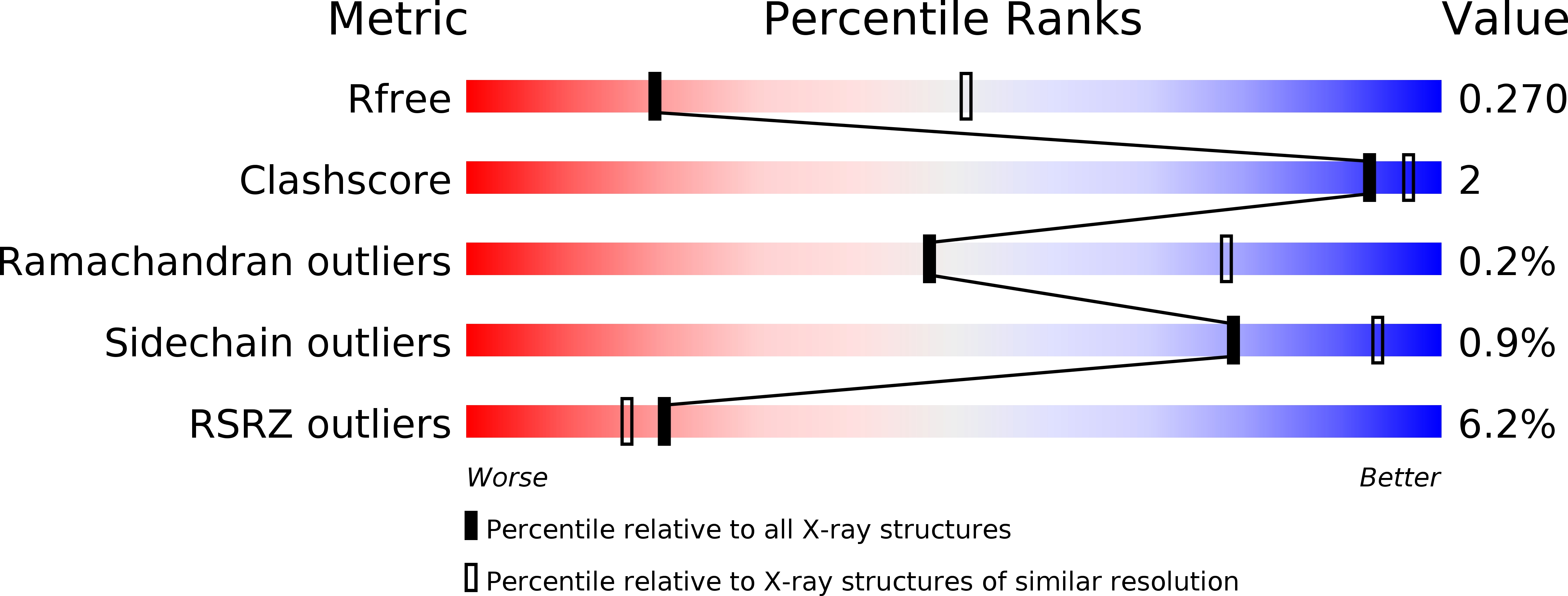
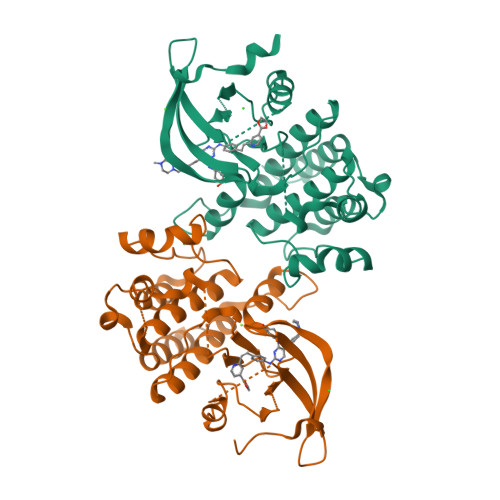
6MEP - PubMed Abstract:
Controlling which particular members of a large protein family are targeted by a drug is key to achieving a desired therapeutic response. In this study, we report a rational data-driven strategy for achieving restricted polypharmacology in the design of antitumor agents selectively targeting the TYRO3, AXL, and MERTK (TAM) family tyrosine kinases. Our computational approach, based on the concept of fragments in structural environments (FRASE), distills relevant chemical information from structural and chemogenomic databases to assemble a three-dimensional inhibitor structure directly in the protein pocket. Target engagement by the inhibitors designed led to disruption of oncogenic phenotypes as demonstrated in enzymatic assays and in a panel of cancer cell lines, including acute lymphoblastic and myeloid leukemia (ALL/AML) and nonsmall cell lung cancer (NSCLC). Structural rationale underlying the approach was corroborated by X-ray crystallography. The lead compound demonstrated potent target inhibition in a pharmacodynamic study in leukemic mice.
Organizational Affiliation:
Center for Integrative Chemical Biology and Drug Discovery, Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy , University of North Carolina at Chapel Hill , Chapel Hill , North Carolina 27599-7363 , United States.