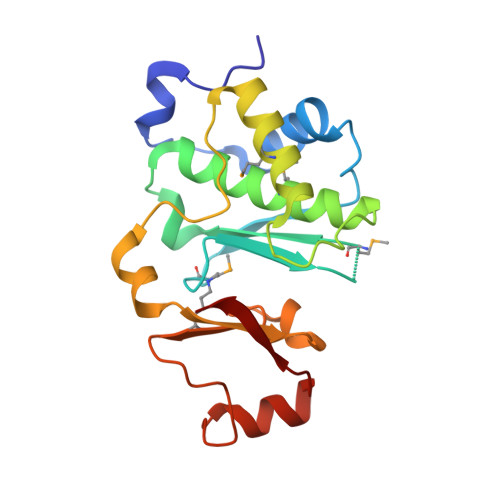
Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan.
Li, F., Raczynska, J.E., Chen, Z., Yu, H.(2019) Cell Rep 26: 3336-3346.e4
- PubMed: 30893605
- DOI: https://doi.org/10.1016/j.celrep.2019.02.082
- Primary Citation of Related Structures:
6MDW, 6MDX - PubMed Abstract:
The DNA-dependent metalloprotease Spartan (SPRTN) cleaves DNA-protein crosslinks (DPCs) and protects cells from DPC-induced genome instability. Germline mutations of SPRTN are linked to human Ruijs-Aalfs syndrome (RJALS) characterized by progeria and early-onset hepatocellular carcinoma. The mechanism of DNA-mediated activation of SPRTN is not understood. Here, we report the crystal structure of the human SPRTN SprT domain bound to single-stranded DNA (ssDNA). Our structure reveals a Zn 2+ -binding sub-domain (ZBD) in SprT that shields its active site located in the metalloprotease sub-domain (MPD). The narrow catalytic groove between MPD and ZBD only permits cleavage of flexible substrates. The ZBD contains an ssDNA-binding site, with a DNA-base-binding pocket formed by aromatic residues. Mutations of ssDNA-binding residues diminish the protease activity of SPRTN. We propose that the ZBD contributes to the ssDNA specificity of SPRTN, restricts the access of globular substrates, and positions DPCs, which may need to be partially unfolded, for optimal cleavage.
Organizational Affiliation:
Department of Pharmacology, Howard Hughes Medical Institute.