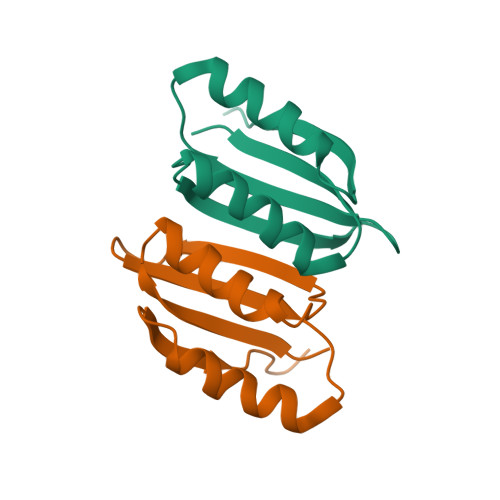
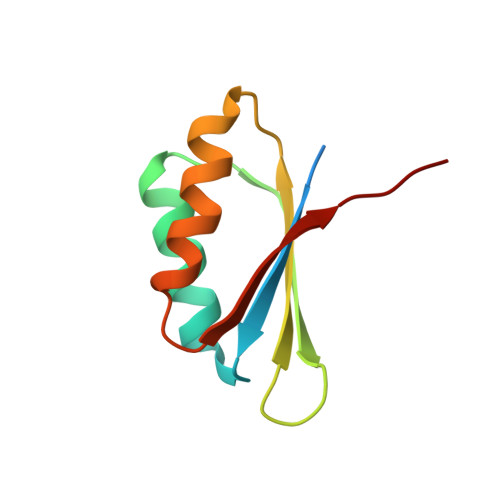
Atomic structure of, and valine binding to the regulatory ACT domain of the Mycobacterium tuberculosis Rel protein.
Shin, J., Singal, B., Sony Subramanian Manimekalai, M., Wei Chen, M., Ragunathan, P., Gruber, G.(2021) FEBS J 288: 2377-2397
- PubMed: 33067840
- DOI: https://doi.org/10.1111/febs.15600
- Primary Citation of Related Structures:
6LXG - PubMed Abstract:
The stringent response, regulated by the bifunctional (p)ppGpp synthetase/hydrolase Rel in mycobacteria, is critical for long-term survival of the drug-tolerant dormant state of Mycobacterium tuberculosis. During amino acid starvation, MtRel senses a drop in amino acid concentration and synthesizes the messengers pppGpp and ppGpp, collectively called (p)ppGpp. Here, we investigate the role of the regulatory 'Aspartokinase, Chorismate mutase and TyrA' (ACT) domain in MtRel. Using NMR spectroscopy approaches, we report the high-resolution structure of dimeric MtRel ACT which selectively binds to valine out of all other branched-chain amino acids tested. A set of MtRel ACT mutants were generated to identify the residues required for maintaining the head-to-tail dimer. Through NMR titrations, we determined the crucial residues for binding of valine and show structural rearrangement of the MtRel ACT dimer in the presence of valine. This study suggests the direct involvement of amino acids in (p)ppGpp accumulation mediated by MtRel independent to interactions with stalled ribosomes. Database Structural data are available in the PDB database under the accession number 6LXG.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, Singapore City, Singapore.