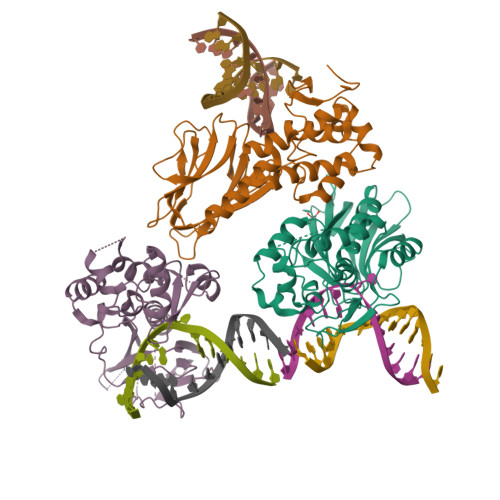
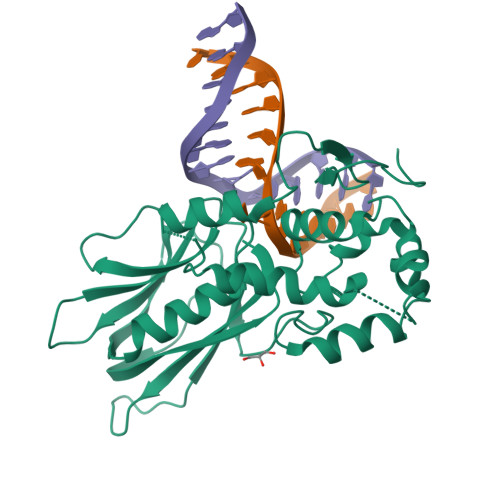
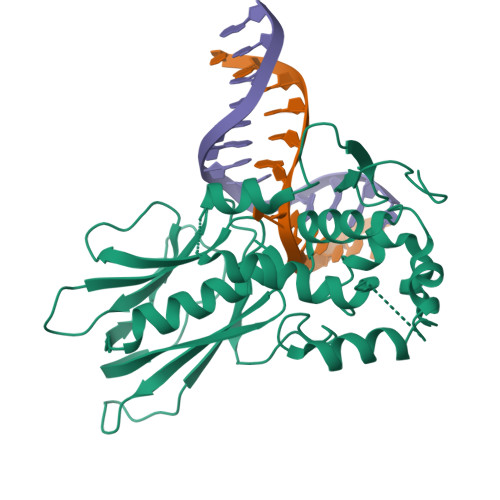
DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Liu, M., Zhang, J., Zhu, C., Zhang, X., Xiao, W., Yan, Y., Liu, L., Zeng, H., Gao, Y.Q., Yi, C.(2021) Nat Commun 12: 4108-4108
- PubMed: 34226550
- DOI: https://doi.org/10.1038/s41467-021-24431-y
- Primary Citation of Related Structures:
6LWA, 6LWB, 6LWC, 6LWD, 6LWF, 6LWG, 6LWH, 6LWI, 6LWJ, 6LWK, 6LWL, 6LWM, 6LWN, 6LWO, 6LWP, 6LWQ, 6LWR - PubMed Abstract:
DNA glycosylases must distinguish the sparse damaged sites from the vast expanse of normal DNA bases. However, our understanding of the nature of nucleobase interrogation is still limited. Here, we show that hNEIL1 (human endonuclease VIII-like 1) captures base lesions via two competing states of interaction: an activated state that commits catalysis and base excision repair, and a quarantine state that temporarily separates and protects the flipped base via auto-inhibition. The relative dominance of the two states depends on key residues of hNEIL1 and chemical properties (e.g. aromaticity and hydrophilicity) of flipped bases. Such a DNA repair mechanism allows hNEIL1 to recognize a broad spectrum of DNA damage while keeps potential gratuitous repair in check. We further reveal the molecular basis of hNEIL1 activity regulation mediated by post-transcriptional modifications and provide an example of how exquisite structural dynamics serves for orchestrated enzyme functions.
Organizational Affiliation:
Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, China.