Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Chen, T., Xiong, M., Zong, X., Ge, Y., Zhang, H., Wang, M., Won Han, G., Yi, C., Ma, L., Ye, R.D., Xu, Y., Zhao, Q., Wu, B.(2020) Nat Commun 11: 1208-1208
- PubMed: 32139677
- DOI: https://doi.org/10.1038/s41467-020-15009-1
- Primary Citation of Related Structures:
6LW5 - PubMed Abstract:
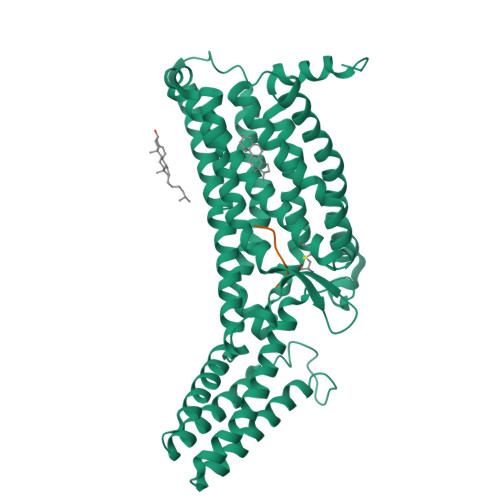
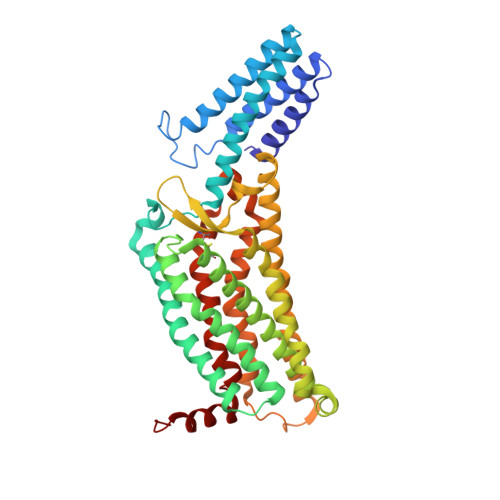
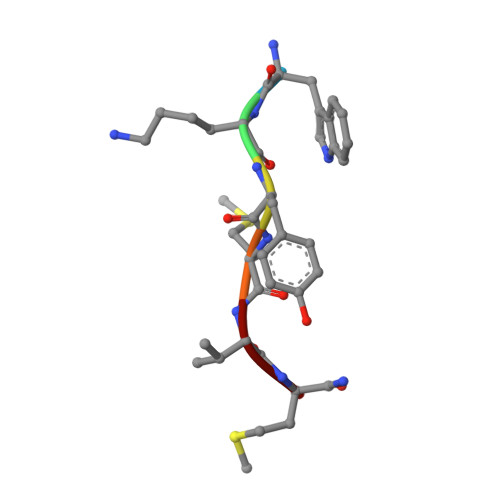
The human formyl peptide receptor 2 (FPR2) plays a crucial role in host defense and inflammation, and has been considered as a drug target for chronic inflammatory diseases. A variety of peptides with different structures and origins have been characterized as FPR2 ligands. However, the ligand-binding modes of FPR2 remain elusive, thereby limiting the development of potential drugs. Here we report the crystal structure of FPR2 bound to the potent peptide agonist WKYMVm at 2.8 Å resolution. The structure adopts an active conformation and exhibits a deep ligand-binding pocket. Combined with mutagenesis, ligand binding and signaling studies, key interactions between the agonist and FPR2 that govern ligand recognition and receptor activation are identified. Furthermore, molecular docking and functional assays reveal key factors that may define binding affinity and agonist potency of formyl peptides. These findings deepen our understanding about ligand recognition and selectivity mechanisms of the formyl peptide receptor family.
Organizational Affiliation:
CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, 201203, Pudong, Shanghai, China.