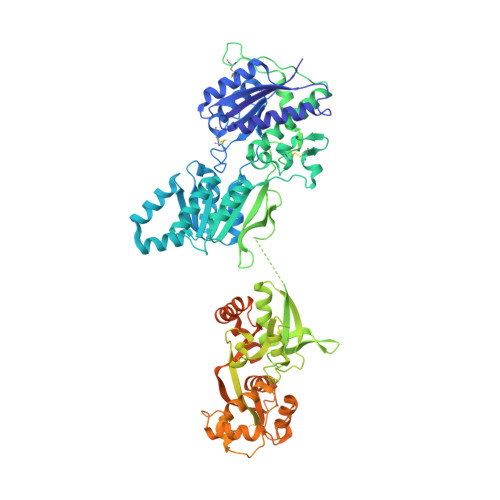
The architecture of GluD2 ionotropic delta glutamate receptor elucidated by cryo-EM.
Burada, A.P., Vinnakota, R., Kumar, J.(2020) J Struct Biol 211: 107546-107546
- PubMed: 32512155
- DOI: https://doi.org/10.1016/j.jsb.2020.107546
- Primary Citation of Related Structures:
6LU9 - PubMed Abstract:
GluD2 receptor belongs to the orphan delta family of glutamate receptor ion channels. These receptors play key roles in synaptogenesis and synaptic plasticity and are associated with multiple neuronal disorders like schizophrenia, autism spectrum disorder, cerebellar ataxia, intellectual disability, paraplegia, retinal dystrophy, etc. Despite the importance of these receptors in CNS, insights into full-length GluD2 receptor structure is missing till-date. Here we report cryo-electron microscopy structure of the rat GluD2 receptor in the presence of calcium ions and the ligand 7-chlorokynurenic acid, elucidating its 3D architecture. The structure reveals a non-swapped architecture at the extracellular amino-terminal (ATD), and ligand-binding domain (LBD) interface similar to that observed in GluD1; however, the organization and arrangement of the ATD and LBD domains in GluD2 are unique. While our results demonstrate that non-swapped architecture is conserved in the delta receptor family, they also highlight the differences that exist between the two member receptors; GluD1 and GluD2.
Organizational Affiliation:
Laboratory of Membrane Protein Biology, National Centre for Cell Science, NCCS Complex, S. P. Pune University, Pune, Maharashtra 411007, India.