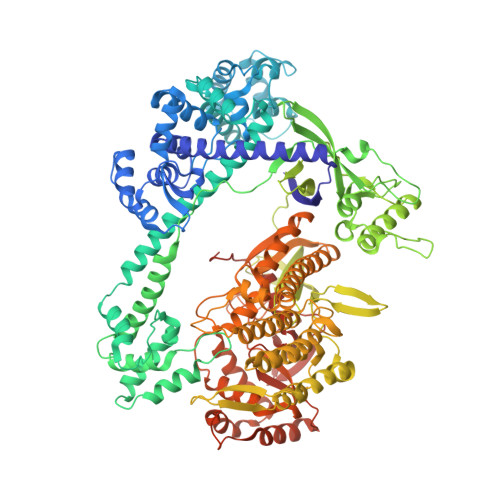
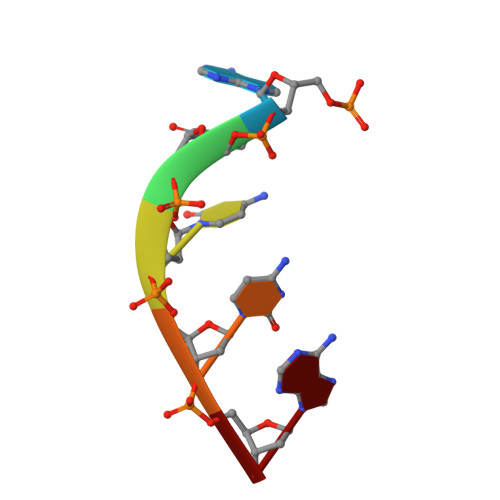
Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Huang, X., Sun, W., Cheng, Z., Chen, M., Li, X., Wang, J., Sheng, G., Gong, W., Wang, Y.(2020) Nat Commun 11: 5241-5241
- PubMed: 33067443
- DOI: https://doi.org/10.1038/s41467-020-19072-6
- Primary Citation of Related Structures:
6LTP, 6LTR, 6LTU, 6LU0 - PubMed Abstract:
To understand how the RuvC catalytic domain of Class 2 Cas proteins cleaves DNA, it will be necessary to elucidate the structures of RuvC-containing Cas complexes in their catalytically competent states. Cas12i2 is a Class 2 type V-I CRISPR-Cas endonuclease that cleaves target dsDNA by an unknown mechanism. Here, we report structures of Cas12i2-crRNA-DNA complexes and a Cas12i2-crRNA complex. We reveal the mechanism of DNA recognition and cleavage by Cas12i2, and activation of the RuvC catalytic pocket induced by a conformational change of the Helical-II domain. The seed region (nucleotides 1-8) is dispensable for RuvC activation, but the duplex of the central spacer (nucleotides 9-15) is required. We captured the catalytic state of Cas12i2, with both metal ions and the ssDNA substrate bound in the RuvC catalytic pocket. Together, our studies provide significant insights into the DNA cleavage mechanism by RuvC-containing Cas proteins.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at the Microscales, University of Science and Technology of China, 230026, Hefei, Anhui, China.