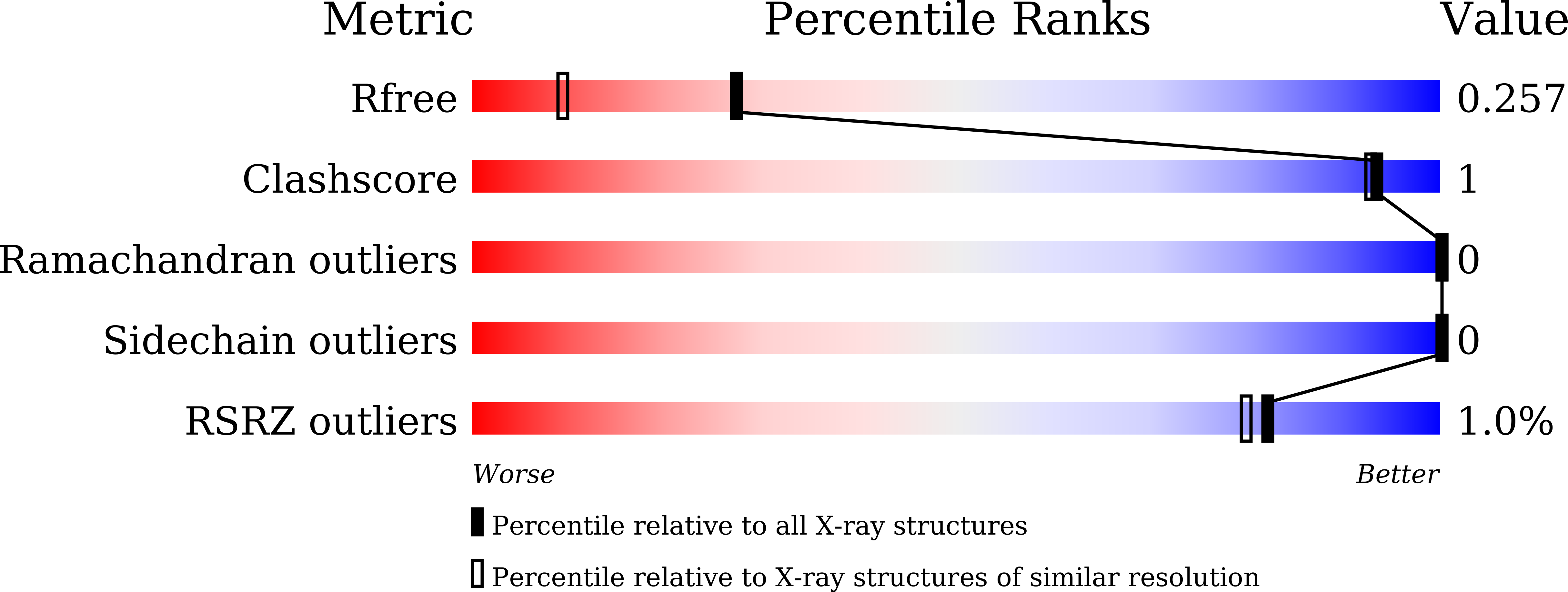
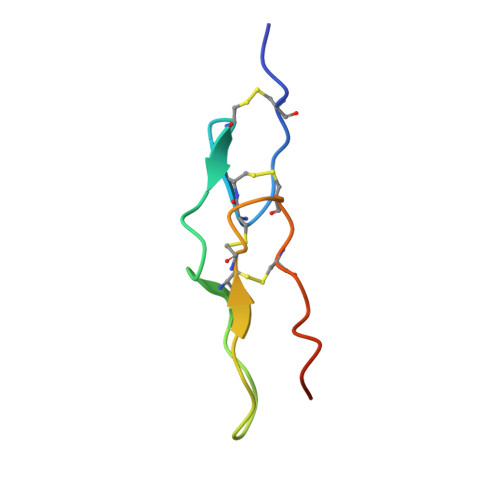
Structural Insight into Integrin Recognition and Anticancer Activity of Echistatin.
Chen, Y.C., Chang, Y.T., Chen, C.Y., Shiu, J.H., Cheng, C.H., Huang, C.H., Chen, J.F., Chuang, W.J.(2020) Toxins (Basel) 12
- PubMed: 33182321
- DOI: https://doi.org/10.3390/toxins12110709
- Primary Citation of Related Structures:
6LSQ - PubMed Abstract:
Echistatin (Ech) is a short disintegrin with a long 42 NPHKGPAT C-terminal tail. We determined the 3-D structure of Ech by X-ray crystallography. Superimposition of the structures of chains A and B showed conformational differences in their RGD loops and C-termini. The chain A structure is consistent with our NMR analysis that the GPAT residues of the C-terminus cannot be observed due to high flexibility. The hydrogen bond patterns of the RGD loop and between the RGD loop and C-terminus in Ech were the same as those of the corresponding residues in medium disintegrins. The mutant with C-terminal HKGPAT truncation caused 6.4-, 7.0-, 11.7-, and 18.6-fold decreases in inhibiting integrins αvβ3, αIIbβ3, αvβ5, and α5β1. Mutagenesis of the C-terminus showed that the H44A mutant caused 2.5- and 4.4-fold increases in inhibiting αIIbβ3 and α5β1, and the K45A mutant caused a 2.6-fold decrease in inhibiting αIIbβ3. We found that Ech inhibited VEGF-induced HUVEC proliferation with an IC 50 value of 103.2 nM and inhibited the migration of A375, U373MG, and Panc-1 tumor cells with IC 50 values of 1.5, 5.7, and 154.5 nM. These findings suggest that Ech is a potential anticancer agent, and its C-terminal region can be optimized to improve its anticancer activity.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, National Cheng Kung University College of Medicine, Tainan 70101, Taiwan.