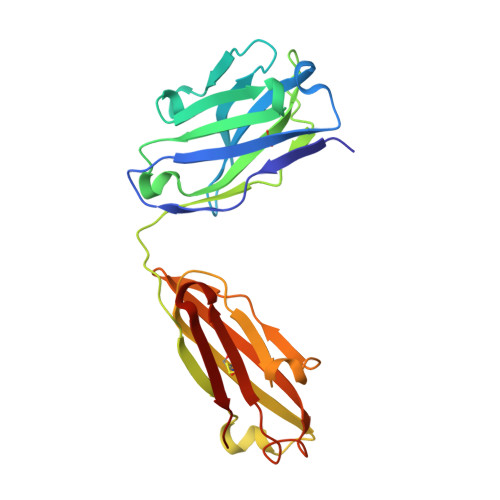
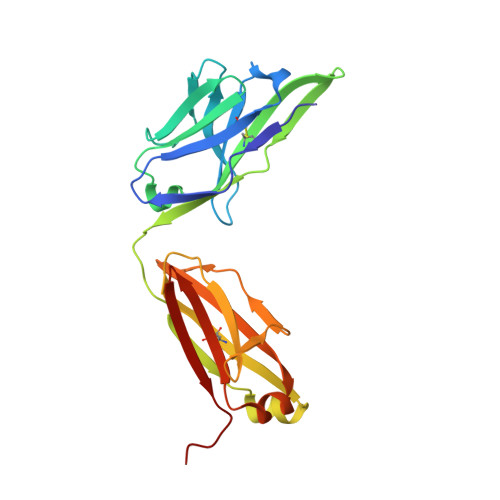
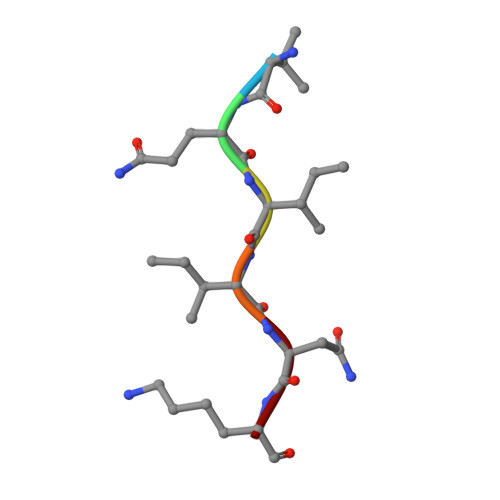
Crystal structure of the human tau PHF core domain VQIINK complexed with the Fab domain of monoclonal antibody Tau2r3.
Tsuchida, T., Susa, K., Kibiki, T., Tsuchiya, T., Miyamoto, K., In, Y., Minoura, K., Taniguchi, T., Ishida, T., Tomoo, K.(2020) FEBS Lett
- PubMed: 32282060
- DOI: https://doi.org/10.1002/1873-3468.13791
- Primary Citation of Related Structures:
6LRA - PubMed Abstract:
Neurofibrillary tangles formed by abnormally aggregated tau protein are a histopathological feature of tauopathies. A tau aggregation inhibitor is a potential therapeutic agent for tauopathies. In this study, we prepared a monoclonal antibody for tau, monoclonal antibody to tau protein (Tau2r3), using as epitope the 272 GGKVQIINKKLD 283 peptide in the microtubule-binding domain of tau, the key region mediating tau aggregation. We show that Tau2r3 clearly inhibits tau aggregation. To analyze the inhibition mechanism of Tau2r3, we solved the crystal structure of the Fab domain of Tau2r3 (Fab2r3) in complex with the VQIINK peptide. In the Fab2r3-VQIINK structure, the second and sixth polar residues and the fourth hydrophobic residue of VQIINK are crucial for binding to Fab2r3. The structural data for the Fab2r3-VQIINK complex could contribute to the design of new tau aggregation inhibitors.
Organizational Affiliation:
Department of Physical Chemistry, Osaka University of Pharmaceutical Sciences, Takatsuki, Japan.