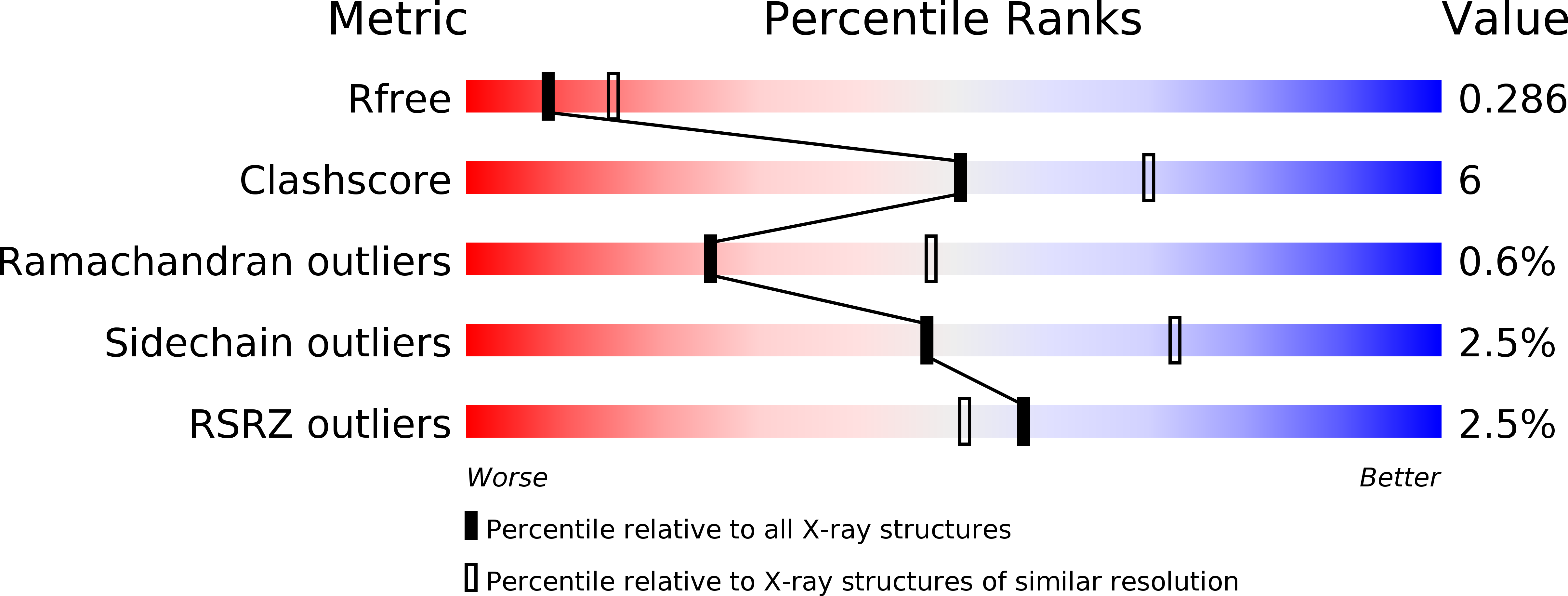
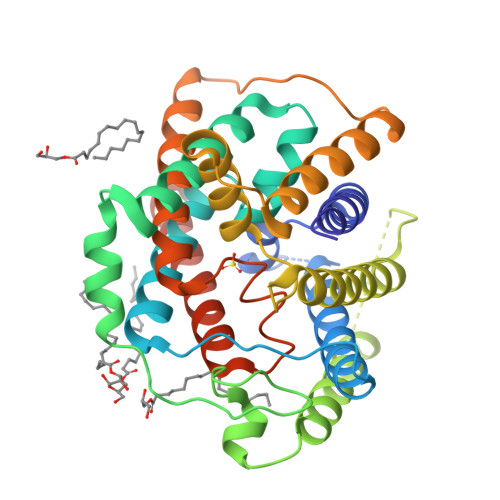
Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Tanaka, Y., Yoshikaie, K., Takeuchi, A., Ichikawa, M., Mori, T., Uchino, S., Sugano, Y., Hakoshima, T., Takagi, H., Nonaka, G., Tsukazaki, T.(2020) Sci Adv 6: eaba7637-eaba7637
- PubMed: 32923628
- DOI: https://doi.org/10.1126/sciadv.aba7637
- Primary Citation of Related Structures:
6LEO, 6LEP - PubMed Abstract:
We have demonstrated that a bacterial membrane protein, YeeE, mediates thiosulfate uptake. Thiosulfate is used for cysteine synthesis in bacteria as an inorganic sulfur source in the global biological sulfur cycle. The crystal structure of YeeE at 2.5-Å resolution reveals an unprecedented hourglass-like architecture with thiosulfate in the positively charged outer concave side. YeeE is composed of loops and 13 helices including 9 transmembrane α helices, most of which show an intramolecular pseudo 222 symmetry. Four characteristic loops are buried toward the center of YeeE and form its central region surrounded by the nine helices. Additional electron density maps and successive molecular dynamics simulations imply that thiosulfate can remain temporally at several positions in the proposed pathway. We propose a plausible mechanism of thiosulfate uptake via three important conserved cysteine residues of the loops along the pathway.
Organizational Affiliation:
Nara Institute of Science and Technology, Ikoma, Nara 630-0192, Japan.