Molecular mechanism of the dual regulatory roles of ATP on the alpha gamma heterodimer of human NAD-dependent isocitrate dehydrogenase.
Sun, P., Bai, T., Ma, T., Ding, J.(2020) Sci Rep 10: 6225-6225
- PubMed: 32277159
- DOI: https://doi.org/10.1038/s41598-020-63425-6
- Primary Citation of Related Structures:
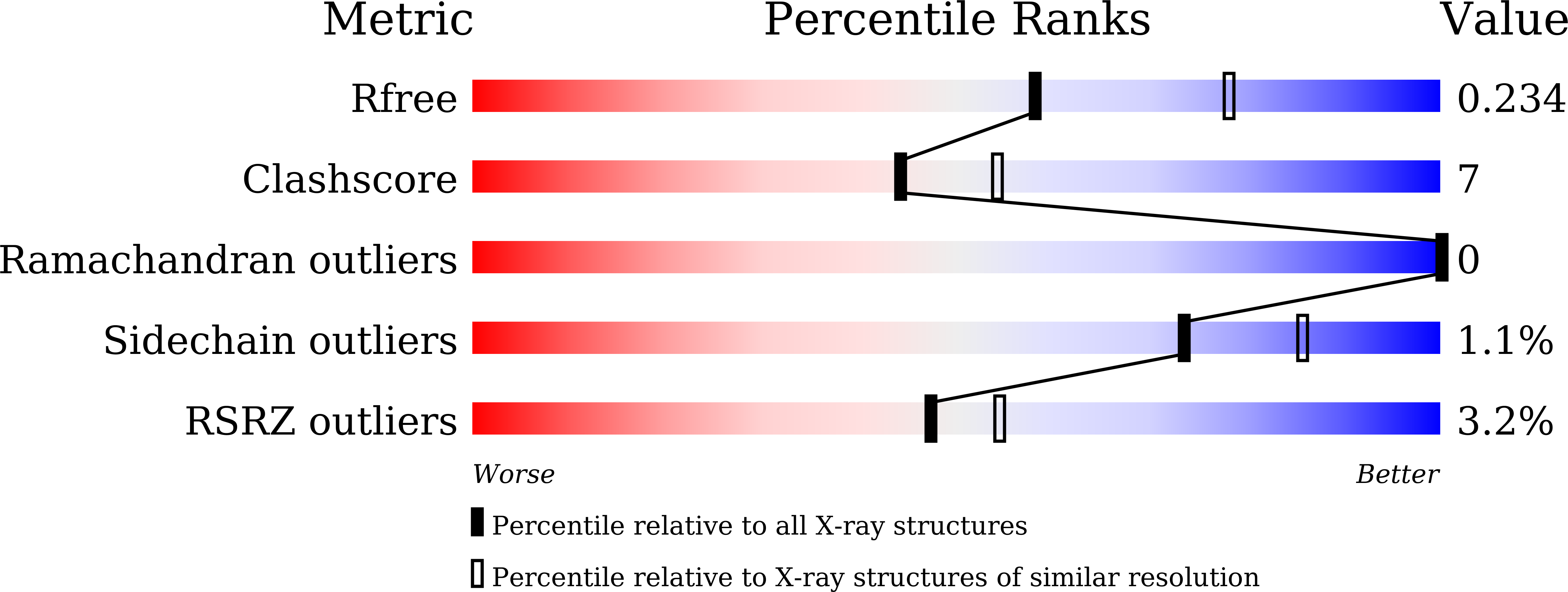
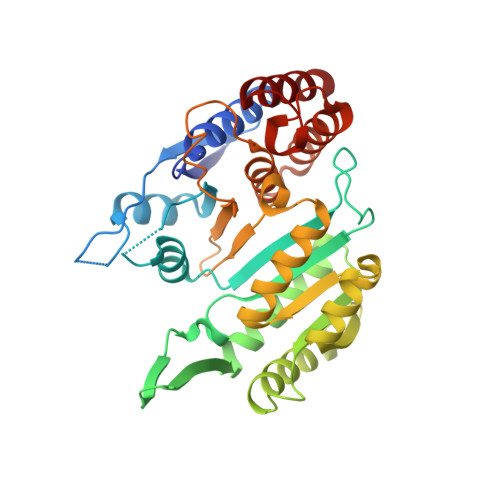
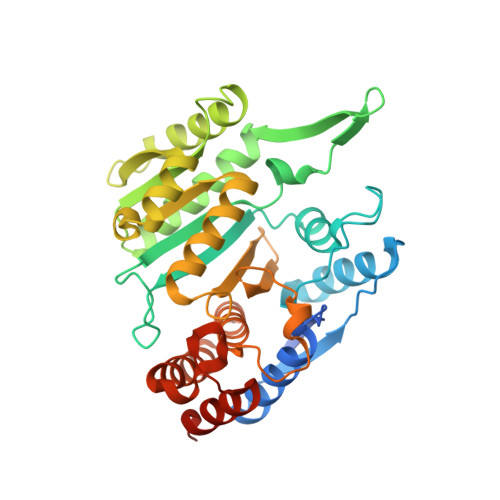
6L57, 6L59 - PubMed Abstract:
Human NAD-dependent isocitrate dehydrogenase (NAD-IDH) is responsible for the catalytic conversion of isocitrate into α-ketoglutarate in the Krebs cycle. This enzyme exists as the α 2 βγ heterotetramer composed of the αβ and αγ heterodimers. Our previous biochemical data showed that the αγ heterodimer and the holoenzyme can be activated by low concentrations of ATP but inhibited by high concentrations of ATP; however, the molecular mechanism was unknown. Here, we report the crystal structures of the αγ heterodimer with ATP binding only to the allosteric site (α Mg γ Mg+CIT+ATP ) and to both the allosteric site and the active site (α Mg+ATP γ Mg+CIT+ATP ). Structural data show that ATP at low concentrations can mimic ADP to bind to the allosteric site, which stabilizes CIT binding and leads the enzyme to adopt an active conformation, revealing why the enzyme can be activated by low concentrations of ATP. On the other hand, at high concentrations ATP is competitive with NAD for binding to the catalytic site. In addition, our biochemical data show that high concentrations of ATP promote the formation of metal ion-ATP chelates. This reduces the concentration of free metal ion available for the catalytic reaction, and thus further inhibits the enzymatic activity. The combination of these two effects accounts for the inhibition of the enzyme at high concentrations of ATP. Taken together, our structural and biochemical data reveal the molecular mechanism for the dual regulatory roles of ATP on the αγ heterodimer of human NAD-IDH.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai, 200031, China.