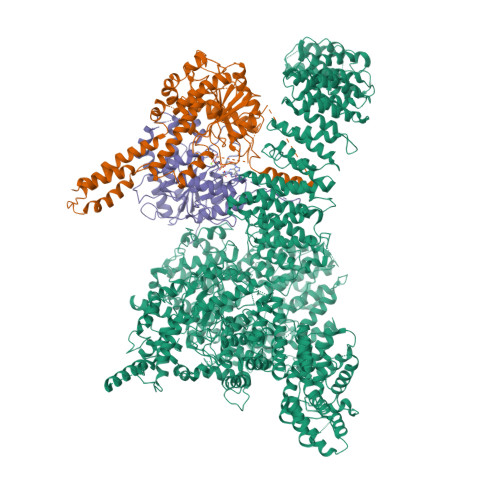
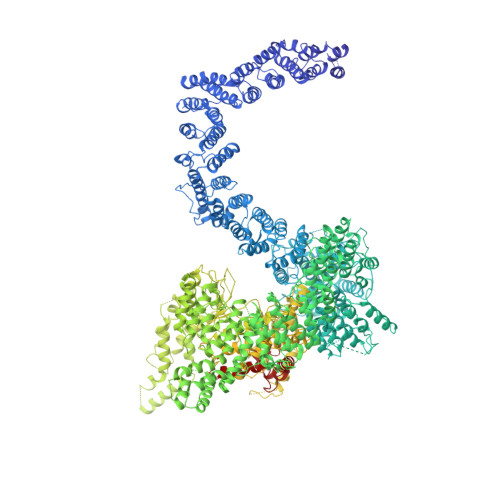
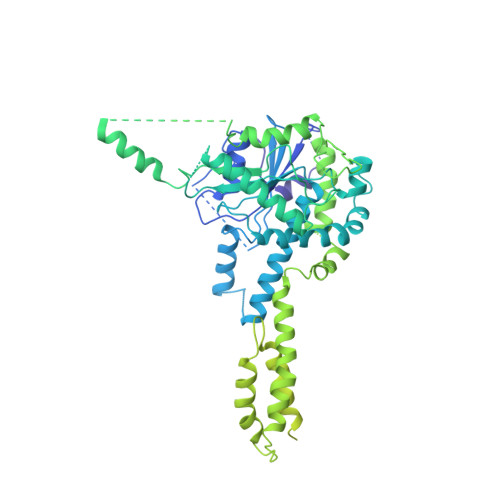
Cryo-EM structure of SMG1-SMG8-SMG9 complex.
Zhu, L., Li, L., Qi, Y., Yu, Z., Xu, Y.(2019) Cell Res 29: 1027-1034
- PubMed: 31729466
- DOI: https://doi.org/10.1038/s41422-019-0255-3
- Primary Citation of Related Structures:
6L53, 6L54 - PubMed Abstract:
Nonsense-mediated mRNA decay (NMD) targets premature stop codon (PTC)-containing mRNAs for rapid degradation, and is essential for mammalian embryonic development, brain development and modulation of the stress response. The key event in NMD is the SMG1-mediated phosphorylation of an RNA helicase UPF1 and SMG1 kinase activity is inhibited by SMG8 and SMG9 in an unknown mechanism. Here, we determined the cryo-EM structures of human SMG1 at 3.6 Å resolution and the SMG1-SMG8-SMG9 complex at 3.4 Å resolution, respectively. SMG8 has a C-terminal kinase inhibitory domain (KID), which covers the catalytic pocket and inhibits the kinase activity of SMG1. Structural analyses suggest that GTP hydrolysis of SMG9 would lead to a dramatic conformational change of SMG8-SMG9 and the KID would move away from the inhibitory position to restore SMG1 kinase activity. Thus, our structural and biochemical analyses provide a mechanistic understanding of SMG1-SMG8-SMG9 complex assembly and the regulatory mechanism of SMG1 kinase activity.
Organizational Affiliation:
Fudan University Shanghai Cancer Center, Institutes of Biomedical Sciences, State Key Laboratory of Genetic Engineering and Key Laboratory of Medical Epigenetics and Metabolism, Shanghai Medical College of Fudan University, Shanghai, 200032, China.