Allosteric Regulation of Hsp90 alpha's Activity by Small Molecules Targeting the Middle Domain of the Chaperone.
Zhou, C., Zhang, C., Zhu, H., Liu, Z., Su, H., Zhang, X., Chen, T., Zhong, Y., Hu, H., Xiong, M., Zhou, H., Xu, Y., Zhang, A., Zhang, N.(2020) iScience 23: 100857-100857
- PubMed: 32058968
- DOI: https://doi.org/10.1016/j.isci.2020.100857
- Primary Citation of Related Structures:
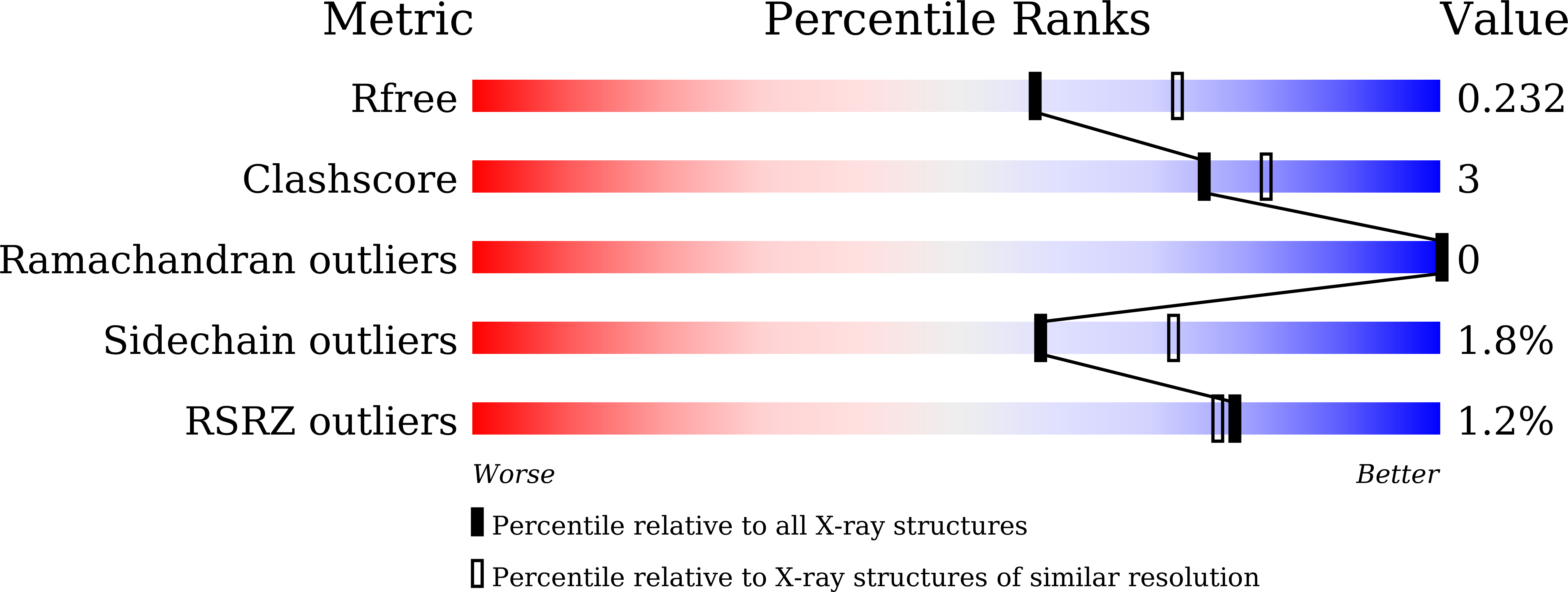
6KSQ - PubMed Abstract:
Hsp90 is a target for anti-cancer drug development. Both the conformational events tuned by ATP/ADP and co-chaperones and the chaperoning cycle timing are required for Hsp90's fully functional display. Interfering with either one of the conformational events or the cycle timing will down-regulate Hsp90's function. In this manuscript, non-covalent allosteric modulators (SOMCL-16-171 and SOMCL-16-175) targeting Hsp90α's middle domain (Hsp90M) were developed for the first time. Multiple techniques were then applied to characterize the interactions between two active compounds and Hsp90α. Two loops and one α-helix (F349-N360, K443-E451, and D372-G387) in Hsp90M were identified responsible for the recognition of SOMCL-16-171 and SOMCL-16-175. Meanwhile, the binding of SOMCL-16-171 and SOMCL-16-175 to Hsp90M was demonstrated to allosterically modulate the structure and function of Hsp90α's N-terminal domain. Finally, cellular assays were conducted to evaluate the cellular activity of SOMCL-16-175, and the results indicate that SOMCL-16-175 destabilizes Hsp90's client proteins and reduces cell viability.
Organizational Affiliation:
Department of Analytical Chemistry, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zu Chong Zhi Road, Shanghai 201203, China.