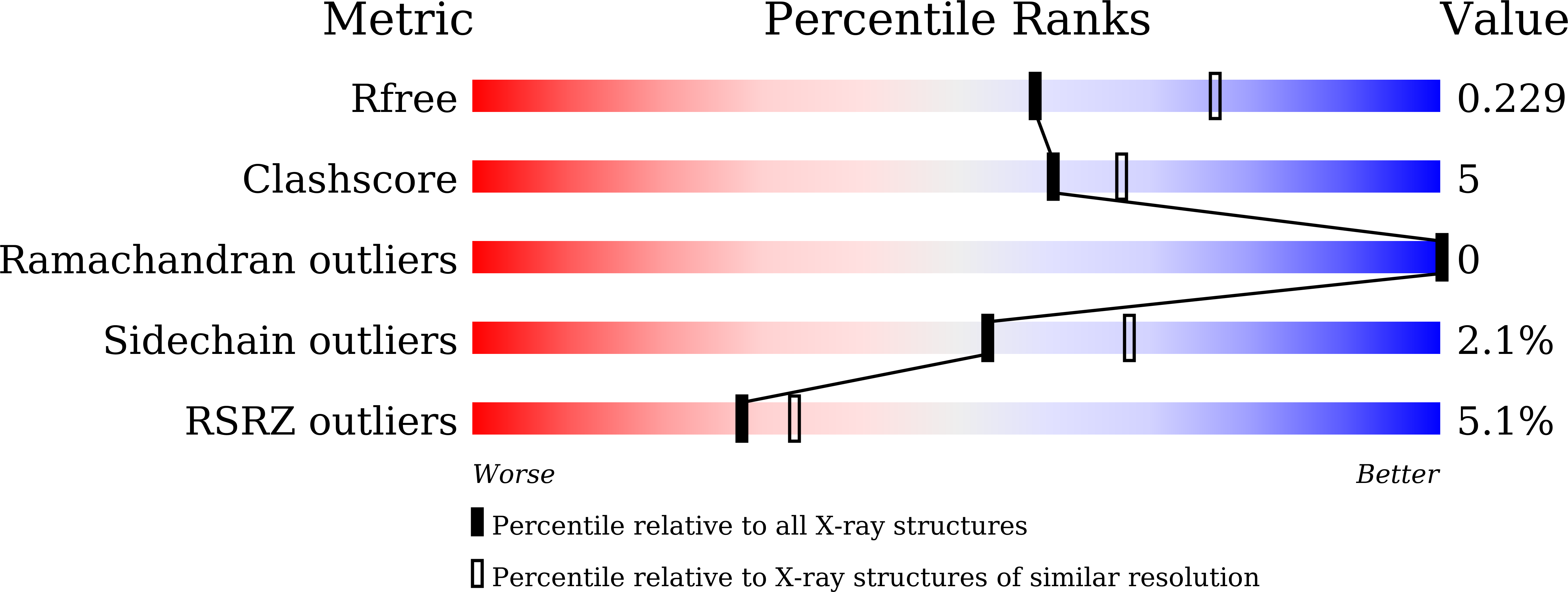
Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Seo, P.W., Jo, H.J., Hwang, I.Y., Jeong, H.Y., Kim, J.H., Kim, J.W., Lee, E.Y., Park, J.B., Kim, J.S.(2020) Catal Sci Technol