Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
Liang, J., Huang, Y.Y., Zhou, Q., Gao, Y., Li, Z., Wu, D., Yu, S., Guo, L., Chen, Z., Huang, L., Liang, S.H., He, X., Wu, R., Luo, H.B.(2020) J Med Chem 63: 3370-3380
- PubMed: 32115956
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00060
- Primary Citation of Related Structures:
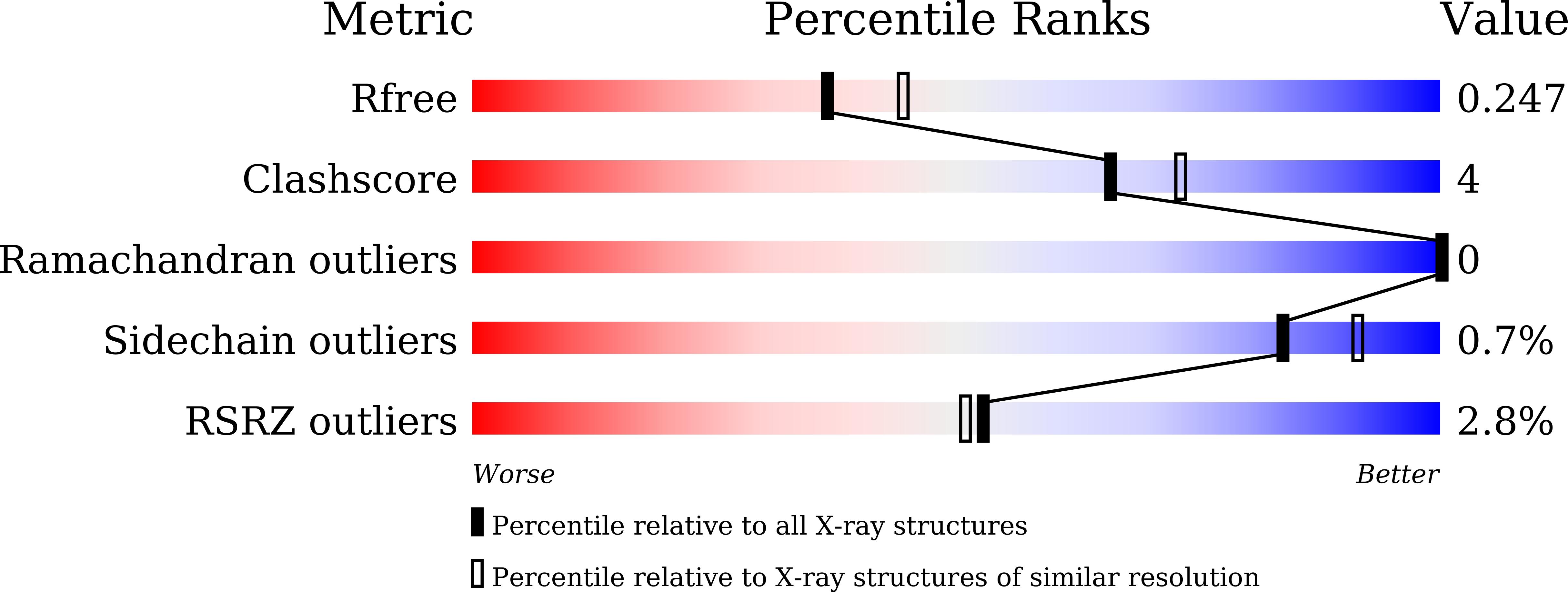
6KJZ, 6KK0 - PubMed Abstract:
To validate PDE4 inhibitors as novel therapeutic agents against vascular dementia (VaD), 25 derivatives were discovered from the natural inhibitor α-mangostin (IC 50 = 1.31 μM). Hit-to-lead optimization identified a novel and selective PDE4 inhibitor 4e (IC 50 = 17 nM), which adopted a different binding pattern from PDE4 inhibitors roflumilast and rolipram. Oral administration of 4e at a dose of 10 mg/kg exhibited remarkable therapeutic effects in a VaD model and did not cause emesis to beagle dogs, indicating its potential as a novel anti-VaD agent.
Organizational Affiliation:
School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou 510006, P. R. China.