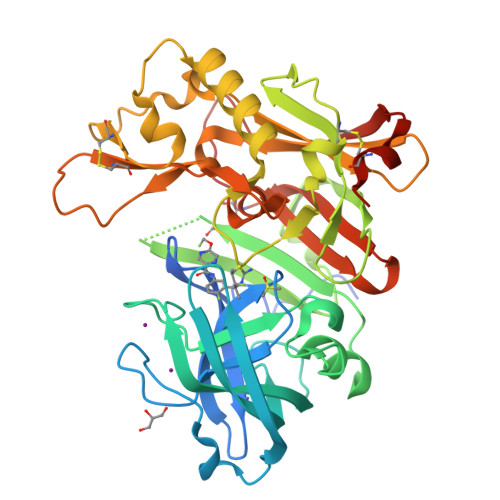
Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
Tadano, G., Komano, K., Yoshida, S., Suzuki, S., Nakahara, K., Fuchino, K., Fujimoto, K., Matsuoka, E., Yamamoto, T., Asada, N., Ito, H., Sakaguchi, G., Kanegawa, N., Kido, Y., Ando, S., Fukushima, T., Teisman, A., Urmaliya, V., Dhuyvetter, D., Borghys, H., Van Den Bergh, A., Austin, N., Gijsen, H.J.M., Yamano, Y., Iso, Y., Kusakabe, K.I.(2019) J Med Chem 62: 9331-9337
- PubMed: 31549838
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01140
- Primary Citation of Related Structures:
6JT3 - PubMed Abstract:
Genetic evidence points to deposition of amyloid-β (Aβ) as a causal factor for Alzheimer's disease. Aβ generation is initiated when β-secretase (BACE1) cleaves the amyloid precursor protein. Starting with an oxazine lead 1 , we describe the discovery of a thiazine-based BACE1 inhibitor 5 with robust Aβ reduction in vivo at low concentrations, leading to a low projected human dose of 14 mg/day where 5 achieved sustained Aβ reduction of 80% at trough level.