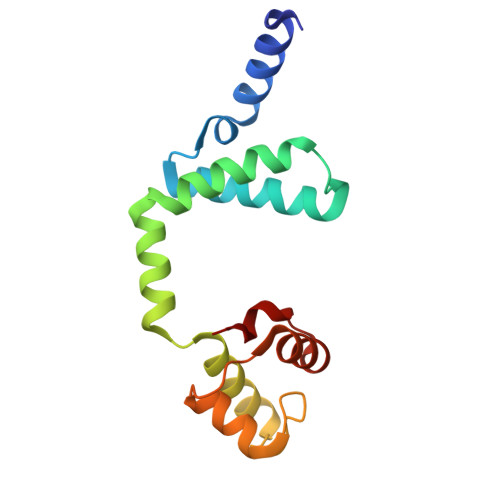
Conformational changes of antitoxin HigA from Escherichia coli str. K-12 upon binding of its cognate toxin HigB reveal a new regulation mechanism in toxin-antitoxin systems.
Xu, B.S., Liu, M., Zhou, K., Geng, Z., Gao, Z.Q., Dong, Y.H., She, Z., Liu, Q.S.(2019) Biochem Biophys Res Commun 514: 37-43
- PubMed: 31014676
- DOI: https://doi.org/10.1016/j.bbrc.2019.04.061
- Primary Citation of Related Structures:
6JQ4 - PubMed Abstract:
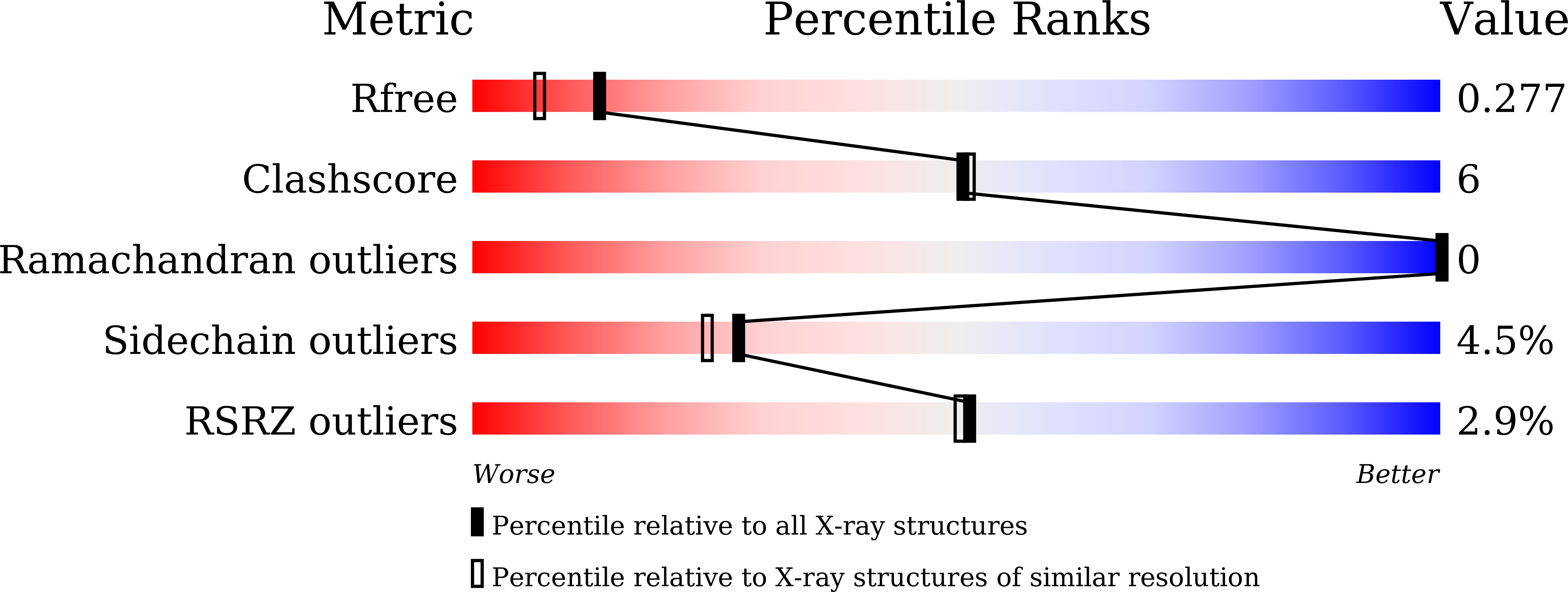
HigA functions as the antitoxin in HigB-HigA toxin-antitoxin system. It neutralizes HigB-mediated toxicity by forming a stable toxin-antitoxin complex. Here the crystal structure of isolated HigA from Escherichia coli str. K-12 has been determined to 2.0 Å resolution. The structural differences between HigA and HigA in HigBA complex imply that HigA undergoes drastic conformational changes upon the binding of HigB. The conformational changes are achieved by rigid motions of N-terminal and C-terminal domains of HigA around its central linker domain, which is different from other known forms of regulation patterns in other organisms. As a transcriptional regulator, HigA bind to its operator DNA through the C-terminal HTH motif, in which key residues were identified in this study.
Organizational Affiliation:
Institute of High Energy Physics, University of Chinese Academy of Sciences, Beijing, 100049, China.