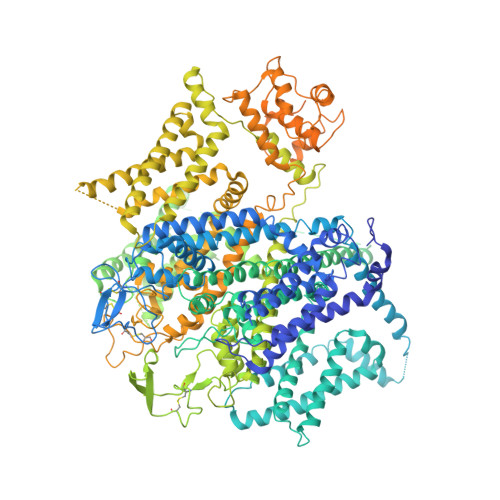
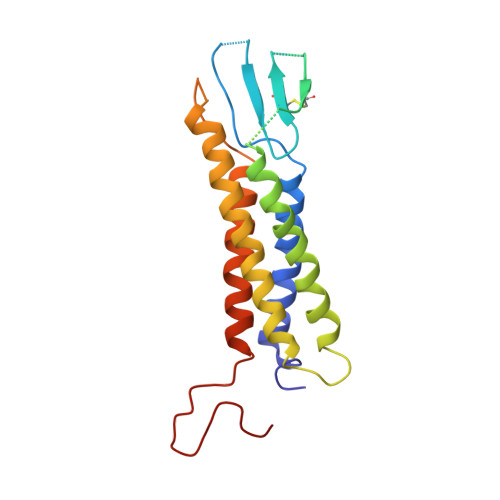
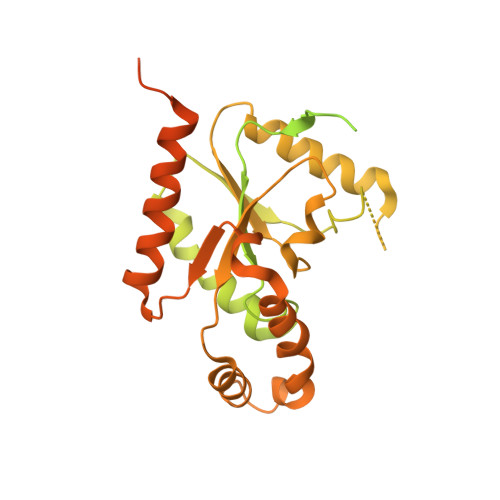
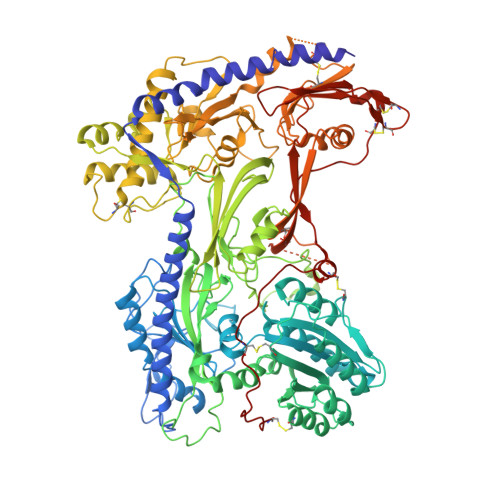
Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Zhao, Y., Huang, G., Wu, J., Wu, Q., Gao, S., Yan, Z., Lei, J., Yan, N.(2019) Cell 177: 1495-1506.e12
- PubMed: 31150622
- DOI: https://doi.org/10.1016/j.cell.2019.04.043
- Primary Citation of Related Structures:
6JP5, 6JP8, 6JPA, 6JPB - PubMed Abstract:
The L-type voltage-gated Ca 2+ (Ca v ) channels are modulated by various compounds exemplified by 1,4-dihydropyridines (DHP), benzothiazepines (BTZ), and phenylalkylamines (PAA), many of which have been used for characterizing channel properties and for treatment of hypertension and other disorders. Here, we report the cryoelectron microscopy (cryo-EM) structures of Ca v 1.1 in complex with archetypal antagonistic drugs, nifedipine, diltiazem, and verapamil, at resolutions of 2.9 Å, 3.0 Å, and 2.7 Å, respectively, and with a DHP agonist Bay K 8644 at 2.8 Å. Diltiazem and verapamil traverse the central cavity of the pore domain, directly blocking ion permeation. Although nifedipine and Bay K 8644 occupy the same fenestration site at the interface of repeats III and IV, the coordination details support previous functional observations that Bay K 8644 is less favored in the inactivated state. These structures elucidate the modes of action of different Ca v ligands and establish a framework for structure-guided drug discovery.
- State Key Laboratory of Membrane Biology, Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: