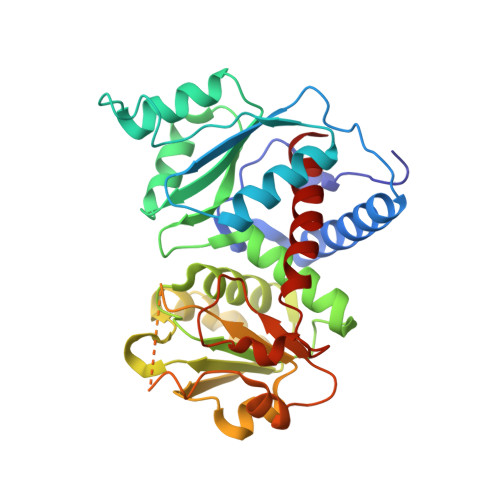
Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase revealed an ordered Bi-Bi reaction mechanism
Matoba, K., Shiba, T., Nara, T., Aoki, T., Nagasaki, S., Hayamizu, R., Honma, T., Tanaka, A., Inoue, M., Matsuoka, S., Balogun, E.O., Inaoka, D.K., Kita, K., Harada, S.To be published.