Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Zhou, Y., Wei, Y., Nanjaraj Urs, A.N., Lin, L., Xu, T., Hu, Y., Ang, E.L., Zhao, H., Yuchi, Z., Zhang, Y.(2019) Biosci Rep 39
- PubMed: 31123167
- DOI: https://doi.org/10.1042/BSR20190715
- Primary Citation of Related Structures:
6JKO, 6JKP - PubMed Abstract:
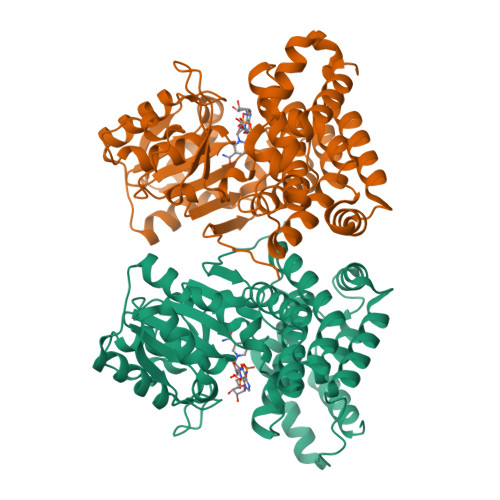
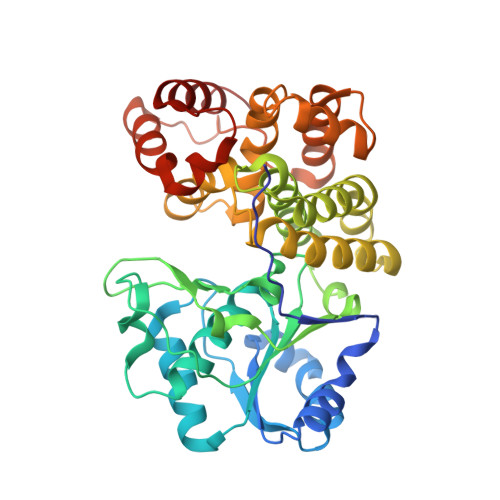
Hydroxyethylsulfonate (isethionate (Ise)) present in mammalian tissues is thought to be derived from aminoethylsulfonate (taurine), as a byproduct of taurine nitrogen assimilation by certain anaerobic bacteria inhabiting the taurine-rich mammalian gut. In previously studied pathways occurring in environmental bacteria, isethionate is generated by the enzyme sulfoacetaldehyde reductase IsfD, belonging to the short-chain dehydrogenase/reductase (SDR) family. An unrelated sulfoacetaldehyde reductase SarD, belonging to the metal-dependent alcohol dehydrogenase superfamily (M-ADH), was recently discovered in the human gut sulfite-reducing bacterium Bilophila wadsworthia ( Bw SarD). Here we report the structural and biochemical characterization of a sulfoacetaldehyde reductase from the human gut fermenting bacterium Bifidobacterium kashiwanohense ( Bk TauF). Bk TauF belongs to the M-ADH family, but is distantly related to Bw SarD (28% sequence identity). The crystal structures of Bk TauF in the apo form and in a binary complex with NAD + were determined at 1.9 and 3.0 Å resolution, respectively. Mutagenesis studies were carried out to investigate the involvement of active site residues in binding the sulfonate substrate. Our studies demonstrate the presence of sulfoacetaldehyde reductase in Bifidobacteria , with a possible role in isethionate production as a byproduct of taurine nitrogen assimilation.
Organizational Affiliation:
Tianjin Key Laboratory for Modern Drug Delivery and High-Efficiency, Collaborative Innovation Center of Chemical Science and Engineering, School of Pharmaceutical Science and Technology, Tianjin University, Tianjin 300072, China.