Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Zhang, Z., Wang, C., Ma, L., Jiang, X., Wu, C., Wang, Y., Jiang, Y., Zheng, W., Yang, Y., Ma, Y., Yang, J.(2019) Biochem Biophys Res Commun 511: 381-386
- PubMed: 30803758
- DOI: https://doi.org/10.1016/j.bbrc.2019.02.064
- Primary Citation of Related Structures:
6JCJ - PubMed Abstract:
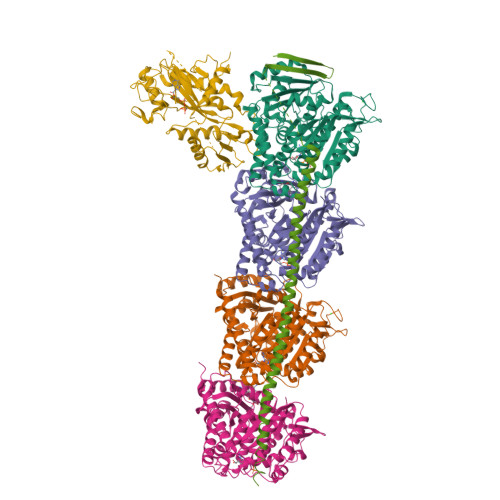
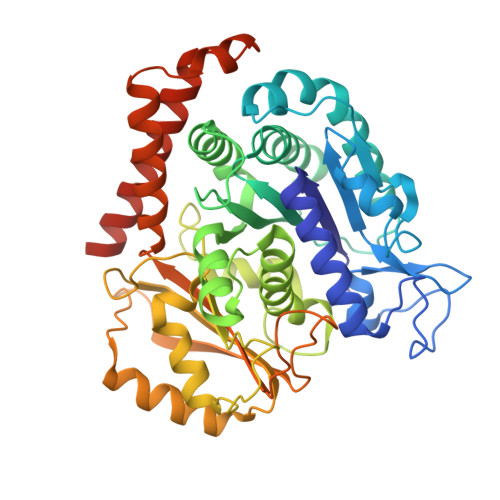
Microtubules (MTs) is one of the most important proteins in eukaryotic cells and plays a key role in the maintenance of cell morphology and cell division. The discovery and development of small molecule drugs targeting MTs has always been an important direction of anti-cancer research. Nowadays 4-Aryl-4H-chromenes have emerged as potent microtubule-targeting agents (MTAs) for various cancers. Crolibulin, a derivative of 4-Aryl-4H-chromenes, which has been progressed to Phase I/II clinical testing's for anaplastic thyroid cancer with the National Cancer Institute. However, the design and development of 4-Aryl-4H-chromenes family drugs have been hindered for a long time by the lack of structural information of the tubulin-agent complex. Here we report a 2.5 Å crystal structure of tubulin complexed with crolibulin. This complex structure reveals the interactions between crolibulin and tubulin, helps explain the results of the structure-activity-relationship (SAR) studies and provides a solid structural basis for the design and development of new 4-Aryl-4H-chromenes derivatives as MTAs.
Organizational Affiliation:
State Key Laboratory of Biotherapy and Cancer Center/Collaborative Innovation Center for Biotherapy, West China Hospital, West China Medical School, Sichuan University, Chengdu, Sichuan, 610041, People's Republic of China. Electronic address: zxzhang1022@163.com.