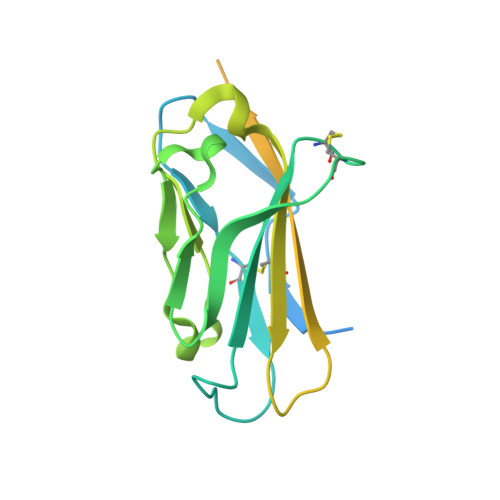
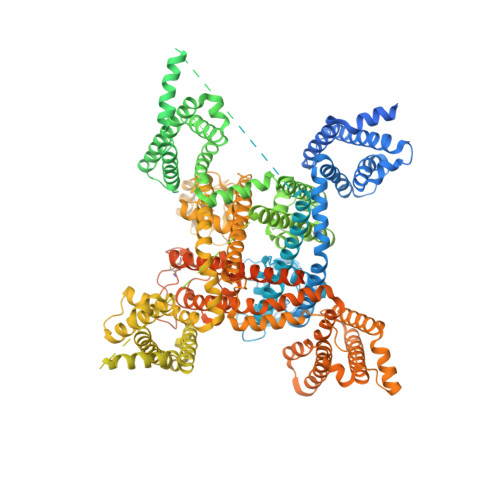
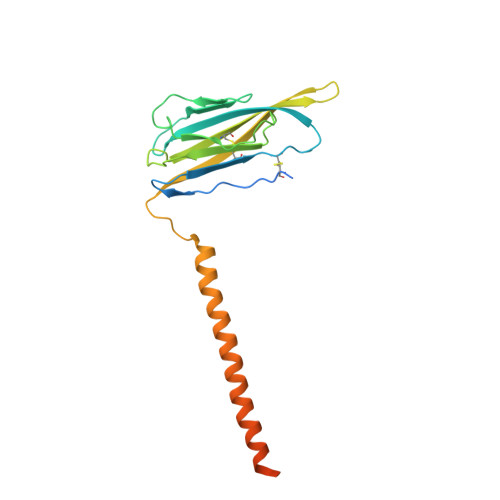
Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Shen, H., Liu, D., Wu, K., Lei, J., Yan, N.(2019) Science 363: 1303-1308
- PubMed: 30765606
- DOI: https://doi.org/10.1126/science.aaw2493
- Primary Citation of Related Structures:
6J8G, 6J8H, 6J8I, 6J8J - PubMed Abstract:
Voltage-gated sodium channel Na v 1.7 represents a promising target for pain relief. Here we report the cryo-electron microscopy structures of the human Na v 1.7-β1-β2 complex bound to two combinations of pore blockers and gating modifier toxins (GMTs), tetrodotoxin with protoxin-II and saxitoxin with huwentoxin-IV, both determined at overall resolutions of 3.2 angstroms. The two structures are nearly identical except for minor shifts of voltage-sensing domain II (VSD II ), whose S3-S4 linker accommodates the two GMTs in a similar manner. One additional protoxin-II sits on top of the S3-S4 linker in VSD IV The structures may represent an inactivated state with all four VSDs "up" and the intracellular gate closed. The structures illuminate the path toward mechanistic understanding of the function and disease of Na v 1.7 and establish the foundation for structure-aided development of analgesics.
- State Key Laboratory of Membrane Biology, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: