Structural basis for activity of TRIC counter-ion channels in calcium release.
Wang, X.H., Su, M., Gao, F., Xie, W., Zeng, Y., Li, D.L., Liu, X.L., Zhao, H., Qin, L., Li, F., Liu, Q., Clarke, O.B., Lam, S.M., Shui, G.H., Hendrickson, W.A., Chen, Y.H.(2019) Proc Natl Acad Sci U S A 116: 4238-4243
- PubMed: 30770441
- DOI: https://doi.org/10.1073/pnas.1817271116
- Primary Citation of Related Structures:
6IYU, 6IYX, 6IYZ, 6IZ0, 6IZ1, 6IZ3, 6IZ4, 6IZ5, 6IZ6, 6IZF - PubMed Abstract:
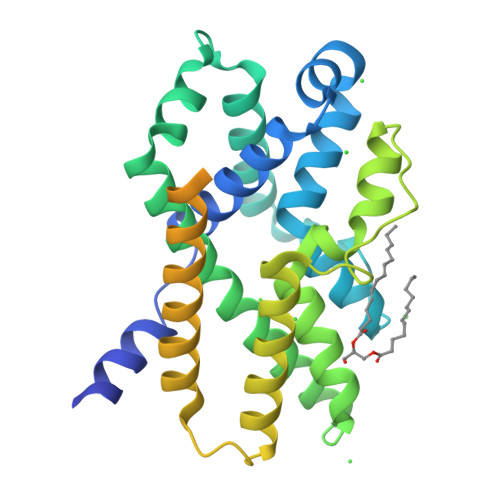
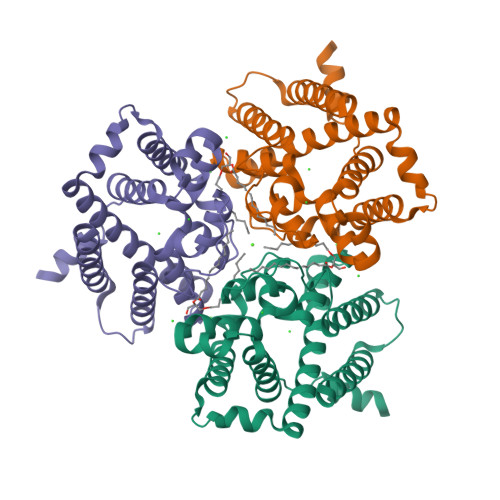
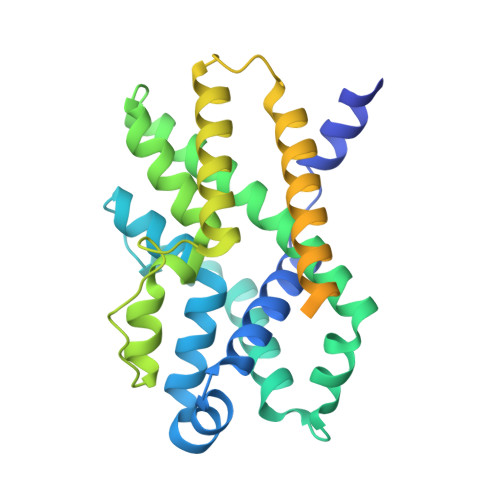
Trimeric intracellular cation (TRIC) channels are thought to provide counter-ion currents that facilitate the active release of Ca 2+ from intracellular stores. TRIC activity is controlled by voltage and Ca 2+ modulation, but underlying mechanisms have remained unknown. Here we describe high-resolution crystal structures of vertebrate TRIC-A and TRIC-B channels, both in Ca 2+ -bound and Ca 2+ -free states, and we analyze conductance properties in structure-inspired mutagenesis experiments. The TRIC channels are symmetric trimers, wherein we find a pore in each protomer that is gated by a highly conserved lysine residue. In the resting state, Ca 2+ binding at the luminal surface of TRIC-A, on its threefold axis, stabilizes lysine blockage of the pores. During active Ca 2+ release, luminal Ca 2+ depletion removes inhibition to permit the lysine-bearing and voltage-sensing helix to move in response to consequent membrane hyperpolarization. Diacylglycerol is found at interprotomer interfaces, suggesting a role in metabolic control.
Organizational Affiliation:
State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.