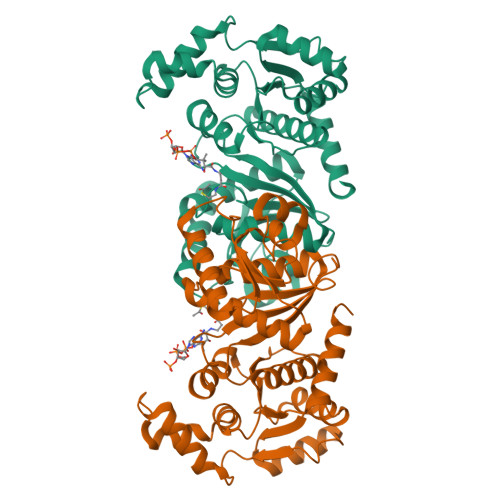
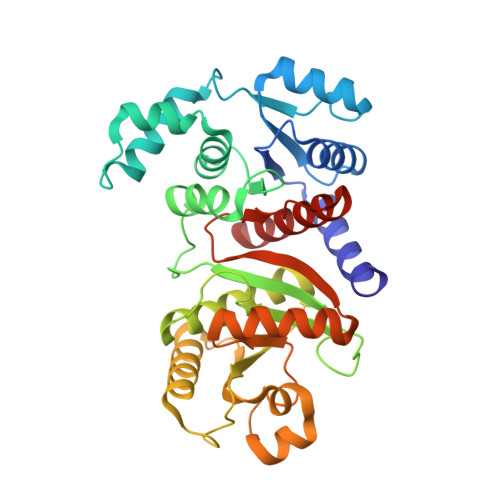
Characterization of the phosphotransacetylase-acetate kinase pathway for ATP production inPorphyromonas gingivalis.
Yoshida, Y., Sato, M., Nonaka, T., Hasegawa, Y., Kezuka, Y.(2019) J Oral Microbiol 11: 1588086-1588086
- PubMed: 31007866
- DOI: https://doi.org/10.1080/20002297.2019.1588086
- Primary Citation of Related Structures:
6IOW, 6IOX, 6IOY - PubMed Abstract:
Acetyl phosphate (AcP) is generally produced from acetyl coenzyme A by phosphotransacetylase (Pta), and subsequent reaction with ADP, catalyzed by acetate kinase (Ack), produces ATP. The mechanism of ATP production in Porphyromonas gingivalis is poorly understood. The aim of this study was to explore the molecular basis of the Pta-Ack pathway in this microorganism. Pta and Ack from P. gingivalis ATCC 33277 were enzymatically and structurally characterized. Structural and mutational analyses suggest that Pta is a dimer with two substrate-binding sites in each subunit. Ack is also dimeric, with a catalytic cleft in each subunit, and structural analysis indicates a dramatic domain motion that opens and closes the cleft during catalysis. ATP formation by Ack proceeds via a sequential mechanism. Reverse transcription-PCR analysis demonstrated that the pta ( PGN_1179 ) and ack ( PGN_1178 ) genes, tandemly located in the genome, are cotranscribed as an operon. Inactivation of pta or ack in P. gingivalis by homologous recombination was successful only when the inactivated gene was expressed in trans . Therefore, both pta and ack genes are essential for this microorganism. Insights into the Pta-Ack pathway reported herein would be helpful to understand the energy acquisition in P. gingivalis .
Organizational Affiliation:
Department of Microbiology, School of Dentistry, Aichi Gakuin University, Nagoya, Japan.