Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Lemaire, S.D., Tedesco, D., Crozet, P., Michelet, L., Fermani, S., Zaffagnini, M., Henri, J.(2018) Antioxidants (Basel) 7
- PubMed: 30477165
- DOI: https://doi.org/10.3390/antiox7120171
- Primary Citation of Related Structures:
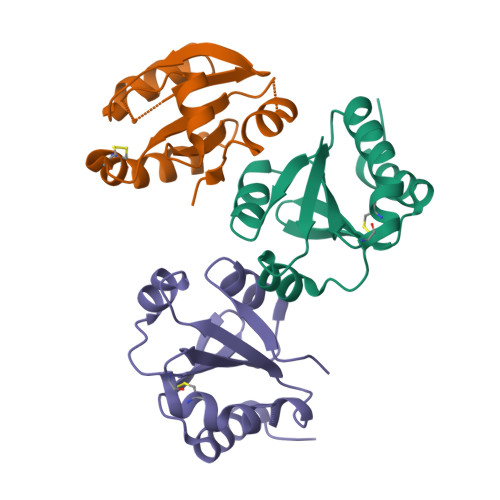
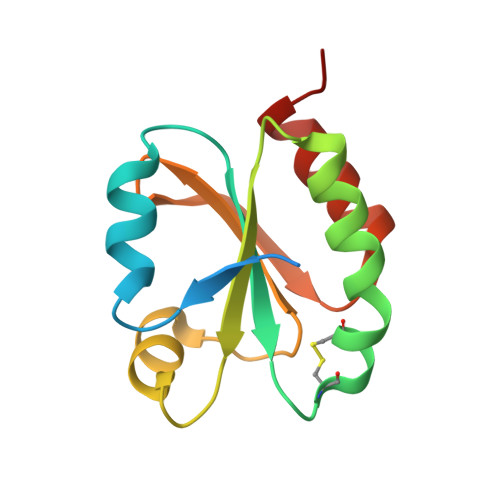
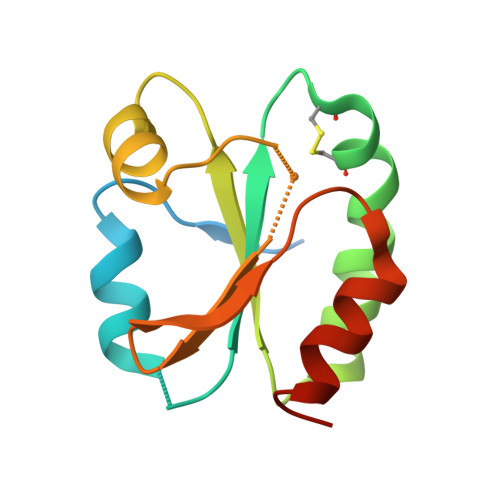
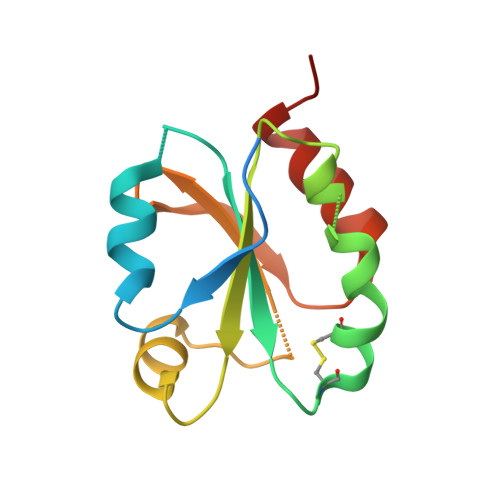
6I19, 6I1C - PubMed Abstract:
Protein disulfide reduction by thioredoxins (TRXs) controls the conformation of enzyme active sites and their multimeric complex formation. TRXs are small oxidoreductases that are broadly conserved in all living organisms. In photosynthetic eukaryotes, TRXs form a large multigenic family, and they have been classified in different types: f, m, x, y, and z types are chloroplastic, while o and h types are located in mitochondria and cytosol. In the model unicellular alga Chlamydomonas reinhardtii , the TRX family contains seven types, with f- and h-types represented by two isozymes. Type-f TRXs interact specifically with targets in the chloroplast, controlling photosynthetic carbon fixation by the Calvin⁻Benson cycle. We solved the crystal structures of TRX f2 and TRX h1 from C. reinhardtii . The systematic comparison of their atomic features revealed a specific conserved electropositive crown around the active site of TRX f, complementary to the electronegative surface of their targets. We postulate that this surface provides specificity to each type of TRX.
Organizational Affiliation:
Laboratoire de Biologie Moléculaire et Cellulaire des Eucaryotes, Institut de Biologie Physico-Chimique, Unité Mixte de Recherche 8226 CNRS Sorbonne Université, 13 rue Pierre et Marie Curie, 75005 Paris, France. lemaire@ibpc.fr.