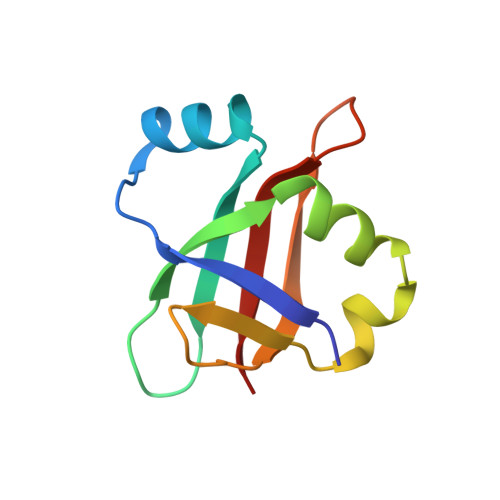
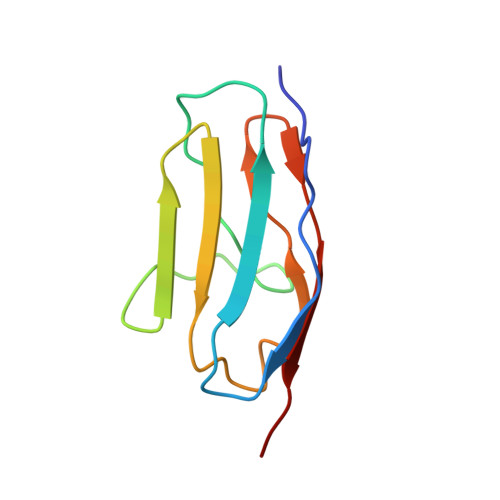
Folding pathway of an Ig domain is conserved on and off the ribosome.
Tian, P., Steward, A., Kudva, R., Su, T., Shilling, P.J., Nickson, A.A., Hollins, J.J., Beckmann, R., von Heijne, G., Clarke, J., Best, R.B.(2018) Proc Natl Acad Sci U S A 115: E11284-E11293
- PubMed: 30413621
- DOI: https://doi.org/10.1073/pnas.1810523115
- Primary Citation of Related Structures:
6I0Y - PubMed Abstract:
Proteins that fold cotranslationally may do so in a restricted configurational space, due to the volume occupied by the ribosome. How does this environment, coupled with the close proximity of the ribosome, affect the folding pathway of a protein? Previous studies have shown that the cotranslational folding process for many proteins, including small, single domains, is directly affected by the ribosome. Here, we investigate the cotranslational folding of an all-β Ig domain, titin I27. Using an arrest peptide-based assay and structural studies by cryo-EM, we show that I27 folds in the mouth of the ribosome exit tunnel. Simulations that use a kinetic model for the force dependence of escape from arrest accurately predict the fraction of folded protein as a function of length. We used these simulations to probe the folding pathway on and off the ribosome. Our simulations-which also reproduce experiments on mutant forms of I27-show that I27 folds, while still sequestered in the mouth of the ribosome exit tunnel, by essentially the same pathway as free I27, with only subtle shifts of critical contacts from the C to the N terminus.
Organizational Affiliation:
Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892.