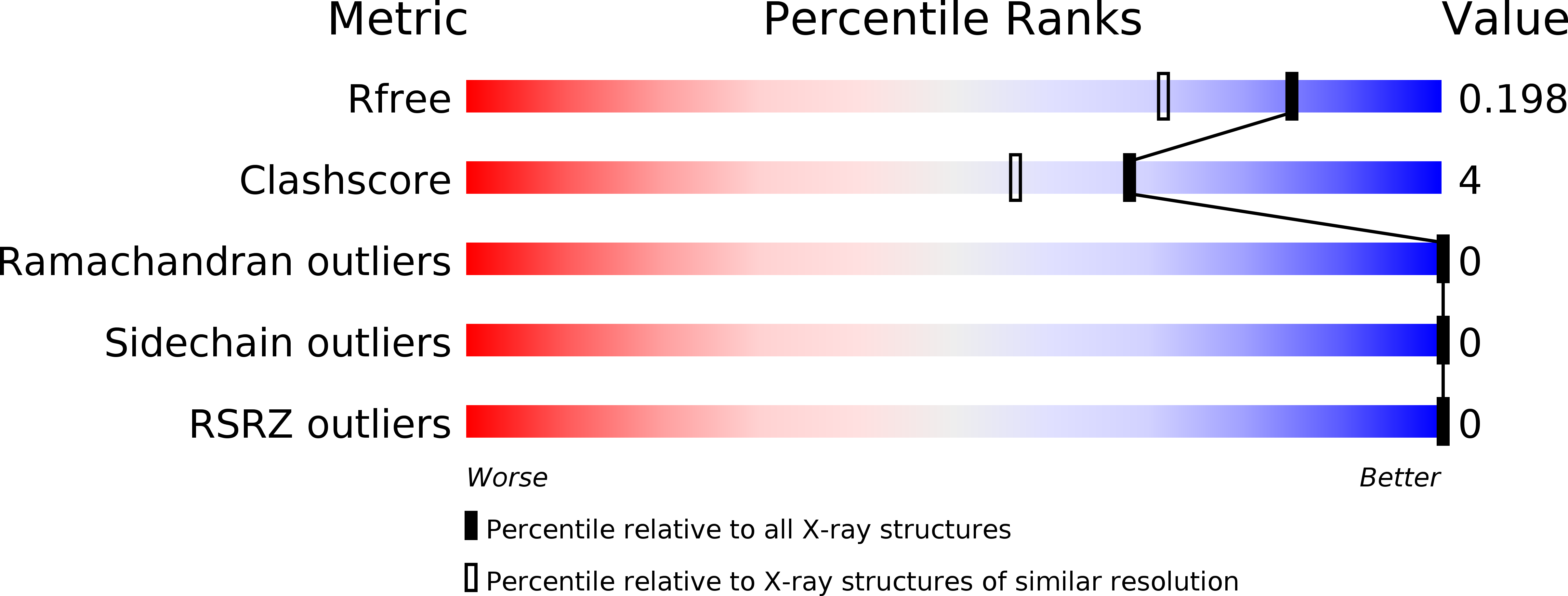
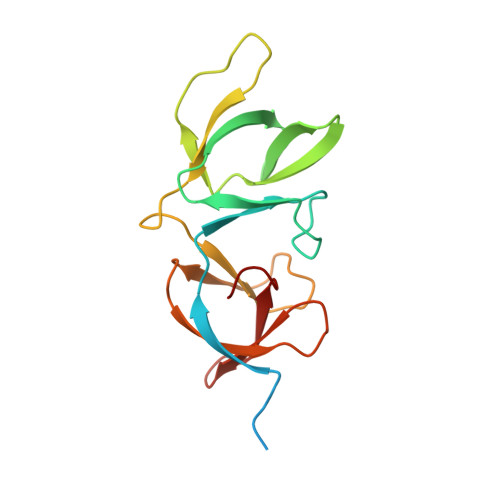
Structural basis for recognition of bacterial cell wall teichoic acid by pseudo-symmetric SH3b-like repeats of a viral peptidoglycan hydrolase
Shen, Y., Kalograiaki, I., Prunotto, A., Dunne, M., Bulous, S., Taylor, N.M., Sumrall, E., Eugster, M.R., Martin, R., Julian-Rodero, A., Gerber, B., Leiman, P.G., Menendez, M., dal Peraro, M., Canada, F.J., Loessner, M.J.(2020) Chem Sci