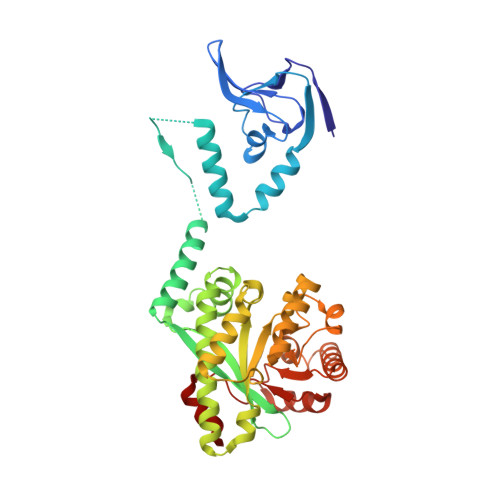
Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Cadby, I.T., Basford, S.M., Nottingham, R., Meek, R., Lowry, R., Lambert, C., Tridgett, M., Till, R., Ahmad, R., Fung, R., Hobley, L., Hughes, W.S., Moynihan, P.J., Sockett, R.E., Lovering, A.L.(2019) EMBO J 38: e100772-e100772
- PubMed: 31355487
- DOI: https://doi.org/10.15252/embj.2018100772
- Primary Citation of Related Structures:
6HQ2, 6HQ3, 6HQ4, 6HQ5, 6HQ7 - PubMed Abstract:
Bacterial usage of the cyclic dinucleotide c-di-GMP is widespread, governing the transition between motile/sessile and unicellular/multicellular behaviors. There is limited information on c-di-GMP metabolism, particularly on regulatory mechanisms governing control of EAL c-di-GMP phosphodiesterases. Herein, we provide high-resolution structures for an EAL enzyme Bd1971, from the predatory bacterium Bdellovibrio bacteriovorus, which is controlled by a second signaling nucleotide, cAMP. The full-length cAMP-bound form reveals the sensory N-terminus to be a domain-swapped variant of the cNMP/CRP family, which in the cAMP-activated state holds the C-terminal EAL enzyme in a phosphodiesterase-active conformation. Using a truncation mutant, we trap both a half-occupied and inactive apo-form of the protein, demonstrating a series of conformational changes that alter juxtaposition of the sensory domains. We show that Bd1971 interacts with several GGDEF proteins (c-di-GMP producers), but mutants of Bd1971 do not share the discrete phenotypes of GGDEF mutants, instead having an elevated level of c-di-GMP, suggesting that the role of Bd1971 is to moderate these levels, allowing "action potentials" to be generated by each GGDEF protein to effect their specific functions.
Organizational Affiliation:
Institute for Microbiology and Infection, School of Biosciences, University of Birmingham, Birmingham, UK.