Characterization of a thermostable flavin-containing monooxygenase from Nitrincola lacisaponensis (NiFMO).
Loncar, N., Fiorentini, F., Bailleul, G., Savino, S., Romero, E., Mattevi, A., Fraaije, M.W.(2019) Appl Microbiol Biotechnol 103: 1755-1764
- PubMed: 30607493
- DOI: https://doi.org/10.1007/s00253-018-09579-w
- Primary Citation of Related Structures:
6HNS - PubMed Abstract:
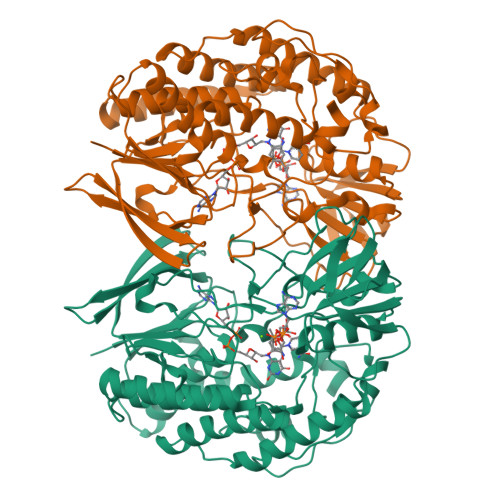
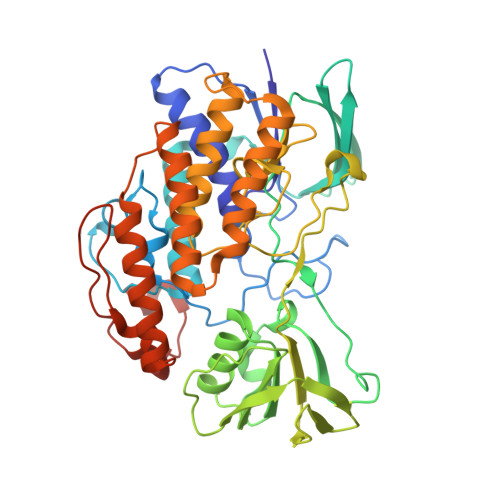
The flavin-containing monooxygenases (FMOs) play an important role in drug metabolism but they also have a high potential in industrial biotransformations. Among the hitherto characterized FMOs, there was no thermostable representative, while such biocatalyst would be valuable for FMO-based applications. Through a targeted genome mining approach, we have identified a gene encoding for a putative FMO from Nitrincola lacisaponensis, an alkaliphilic extremophile bacterium. Herein, we report the biochemical and structural characterization of this newly discovered bacterial FMO (NiFMO). NiFMO can be expressed as active and soluble enzyme at high level in Escherichia coli (90-100 mg/L of culture). NiFMO is relatively thermostable (melting temperature (T m ) of 51 °C), displays high organic solvent tolerance, and accepts a broad range of substrates. The crystal structure of NiFMO was solved at 1.8 Å resolution, which allows future structure-based enzyme engineering. Altogether, NiFMO represents an interesting newly discovered enzyme with the appropriate features to develop into an industrially applied biocatalyst.
Organizational Affiliation:
Groningen Enzyme and Cofactor Collection (GECCO), University of Groningen, Nijenborgh 4, 9747AG, Groningen, The Netherlands.