Structure ofMycobacterium tuberculosisphosphatidylinositol phosphate synthase reveals mechanism of substrate binding and metal catalysis.
Grave, K., Bennett, M.D., Hogbom, M.(2019) Commun Biol 2: 175-175
- PubMed: 31098408
- DOI: https://doi.org/10.1038/s42003-019-0427-1
- Primary Citation of Related Structures:
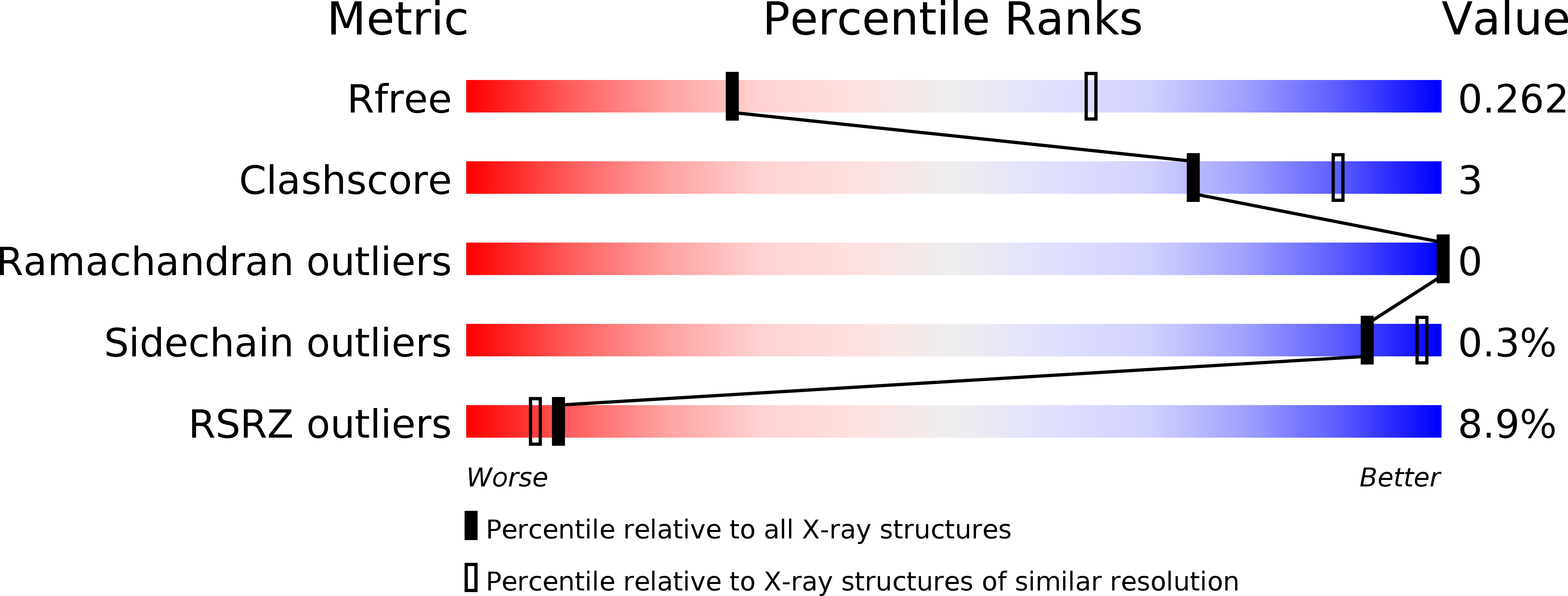
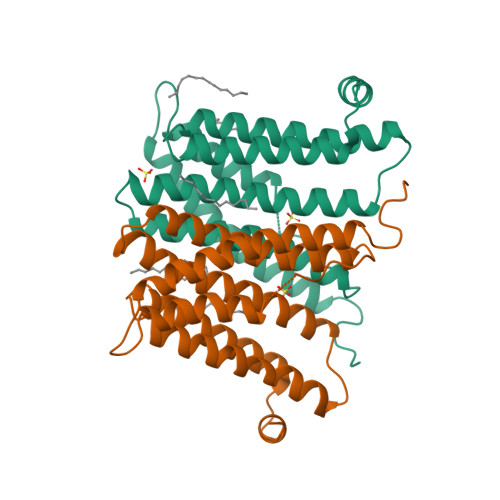
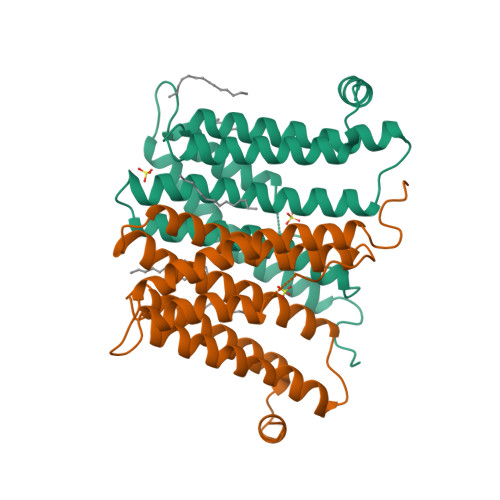
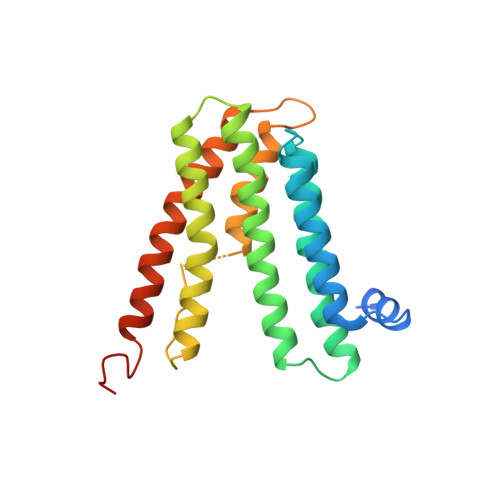
6H53, 6H59, 6H5A - PubMed Abstract:
Tuberculosis causes over one million yearly deaths, and drug resistance is rapidly developing. Mycobacterium tuberculosis phosphatidylinositol phosphate synthase (PgsA1) is an integral membrane enzyme involved in biosynthesis of inositol-derived phospholipids required for formation of the mycobacterial cell wall, and a potential drug target. Here we present three crystal structures of M. tuberculosis PgsA1: in absence of substrates (2.9 Å), in complex with Mn 2+ and citrate (1.9 Å), and with the CDP-DAG substrate (1.8 Å). The structures reveal atomic details of substrate binding as well as coordination and dynamics of the catalytic metal site. In addition, molecular docking supported by mutagenesis indicate a binding mode for the second substrate, D- myo -inositol-3-phosphate. Together, the data describe the structural basis for M. tuberculosis phosphatidylinositol phosphate synthesis and suggest a refined general catalytic mechanism-including a substrate-induced carboxylate shift-for Class I CDP-alcohol phosphotransferases, enzymes essential for phospholipid biosynthesis in all domains of life.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Svante Arrhenius väg 16 C, SE-106 91 Stockholm, Sweden.