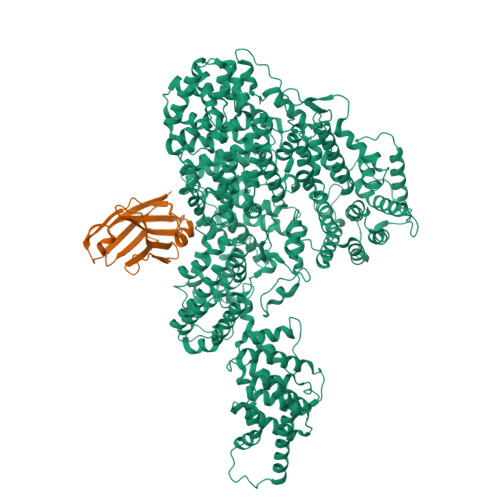
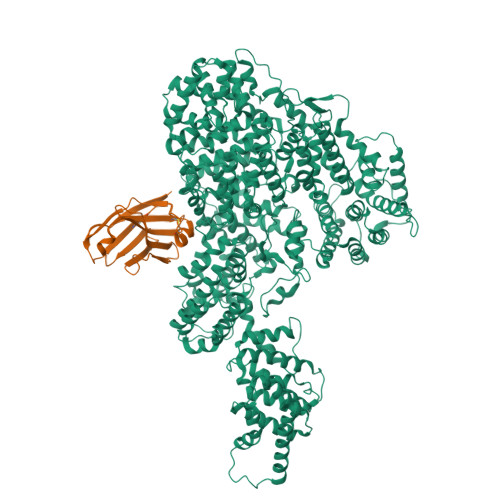
Crystal structure of human Mediator subunit MED23.
Monte, D., Clantin, B., Dewitte, F., Lens, Z., Rucktooa, P., Pardon, E., Steyaert, J., Verger, A., Villeret, V.(2018) Nat Commun 9: 3389-3389
- PubMed: 30140054
- DOI: https://doi.org/10.1038/s41467-018-05967-y
- Primary Citation of Related Structures:
6H02 - PubMed Abstract:
The Mediator complex transduces regulatory information from enhancers to promoters and performs essential roles in the initiation of transcription in eukaryotes. Human Mediator comprises 26 subunits forming three modules termed Head, Middle and Tail. Here we present the 2.8 Å crystal structure of MED23, the largest subunit from the human Tail module. The structure identifies 25 HEAT repeats-like motifs organized into 5 α-solenoids. MED23 adopts an arch-shaped conformation, with an N-terminal domain (Nter) protruding from a large core region. In the core four solenoids, motifs wrap on themselves, creating triangular-shaped structural motifs on both faces of the arch, with extended grooves propagating through the interfaces between the solenoid motifs. MED23 is known to interact with several specific transcription activators and is involved in splicing, elongation, and post-transcriptional events. The structure rationalizes previous biochemical observations and paves the way for improved understanding of the cross-talk between Mediator and transcriptional activators.
Organizational Affiliation:
CNRS, UMR 8576-UGSF- Unité de Glycobiologie Structurale et Fonctionnelle, Univ. Lille, 59000, Lille, France. didier.monte@univ-lille.fr.