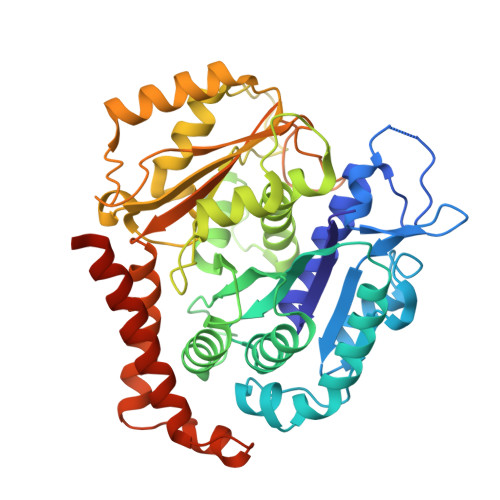
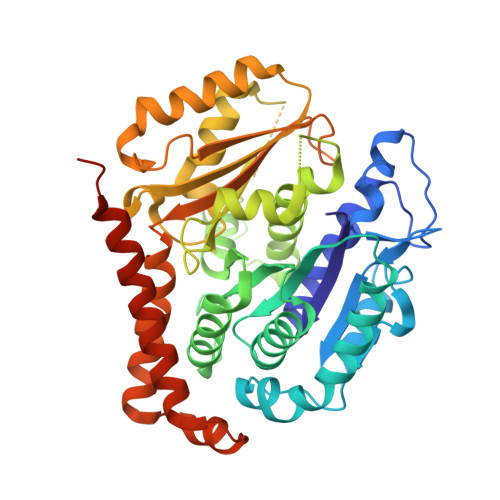
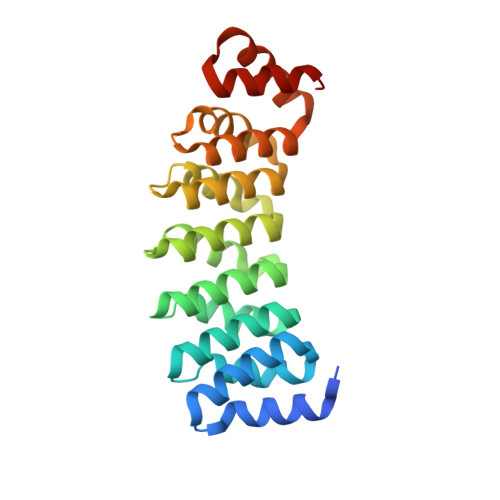
Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Campanacci, V., Urvoas, A., Consolati, T., Cantos-Fernandes, S., Aumont-Nicaise, M., Valerio-Lepiniec, M., Surrey, T., Minard, P., Gigant, B.(2019) Structure 27: 497-506.e4
- PubMed: 30661854
- DOI: https://doi.org/10.1016/j.str.2018.12.001
- Primary Citation of Related Structures:
6GWC, 6GWD - PubMed Abstract:
Microtubules are cytoskeletal filaments of eukaryotic cells made of αβ-tubulin heterodimers. Structural studies of non-microtubular tubulin rely mainly on molecules that prevent its self-assembly and are used as crystallization chaperones. Here we identified artificial proteins from an αRep library that are specific to α-tubulin. Turbidity experiments indicate that these αReps impede microtubule assembly in a dose-dependent manner and total internal reflection fluorescence microscopy further shows that they specifically block growth at the microtubule (-) end. Structural data indicate that they do so by targeting the α-tubulin longitudinal surface. Interestingly, in one of the complexes studied, the α subunit is in a conformation that is intermediate between the ones most commonly observed in X-ray structures of tubulin and those seen in the microtubule, emphasizing the plasticity of tubulin. These α-tubulin-specific αReps broaden the range of tools available for the mechanistic study of microtubule dynamics and its regulation.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ. Paris-Sud, Université Paris-Saclay, Gif-sur-Yvette Cedex 91198, France.