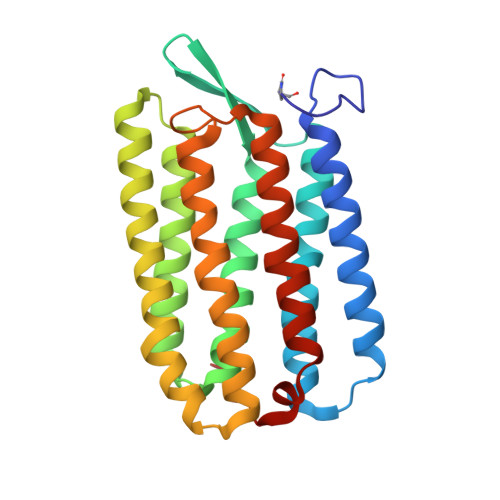
Structures of the archaerhodopsin-3 transporter reveal that disordering of internal water networks underpins receptor sensitization.
Bada Juarez, J.F., Judge, P.J., Adam, S., Axford, D., Vinals, J., Birch, J., Kwan, T.O.C., Hoi, K.K., Yen, H.Y., Vial, A., Milhiet, P.E., Robinson, C.V., Schapiro, I., Moraes, I., Watts, A.(2021) Nat Commun 12: 629-629
- PubMed: 33504778
- DOI: https://doi.org/10.1038/s41467-020-20596-0
- Primary Citation of Related Structures:
6GUX, 6S6C - PubMed Abstract:
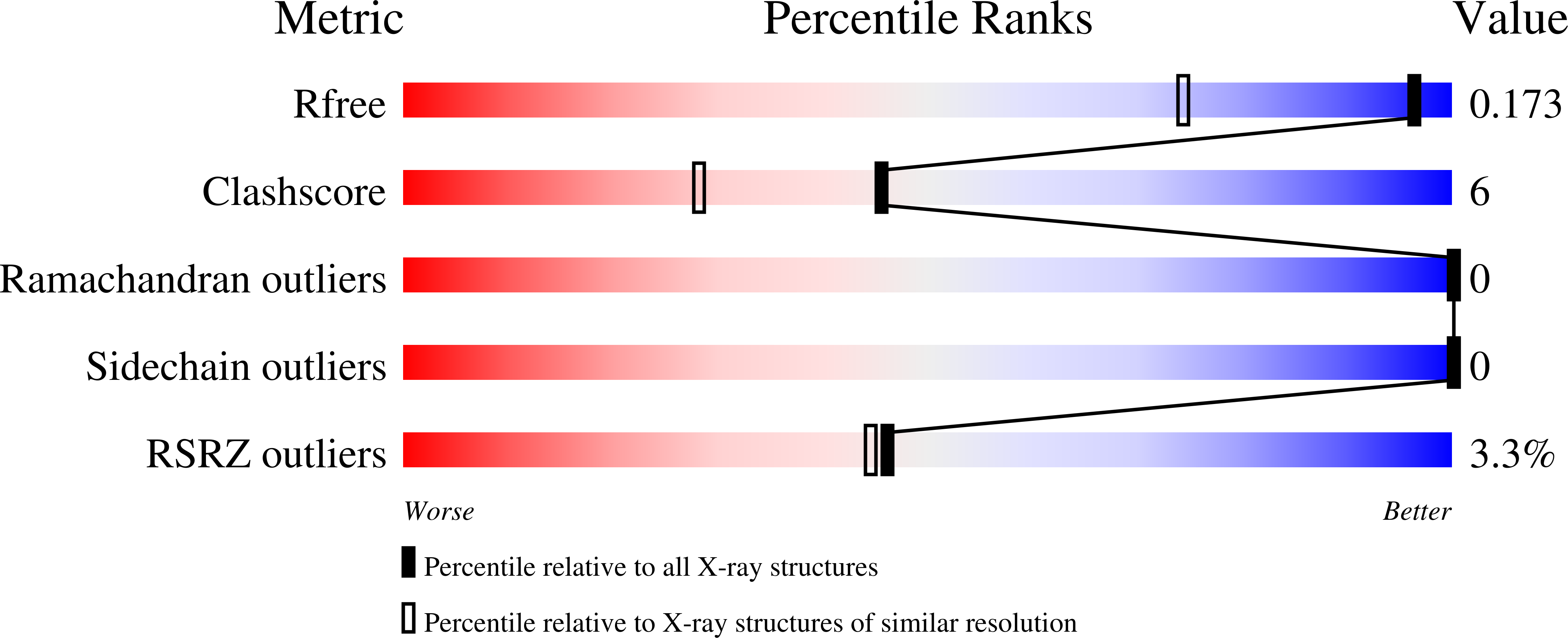
Many transmembrane receptors have a desensitized state, in which they are unable to respond to external stimuli. The family of microbial rhodopsin proteins includes one such group of receptors, whose inactive or dark-adapted (DA) state is established in the prolonged absence of light. Here, we present high-resolution crystal structures of the ground (light-adapted) and DA states of Archaerhodopsin-3 (AR3), solved to 1.1 Å and 1.3 Å resolution respectively. We observe significant differences between the two states in the dynamics of water molecules that are coupled via H-bonds to the retinal Schiff Base. Supporting QM/MM calculations reveal how the DA state permits a thermodynamic equilibrium between retinal isomers to be established, and how this same change is prevented in the ground state in the absence of light. We suggest that the different arrangement of internal water networks in AR3 is responsible for the faster photocycle kinetics compared to homologs.
Organizational Affiliation:
Biochemistry Department, Oxford University, South Parks Road, Oxford, OX1 3QU, UK.