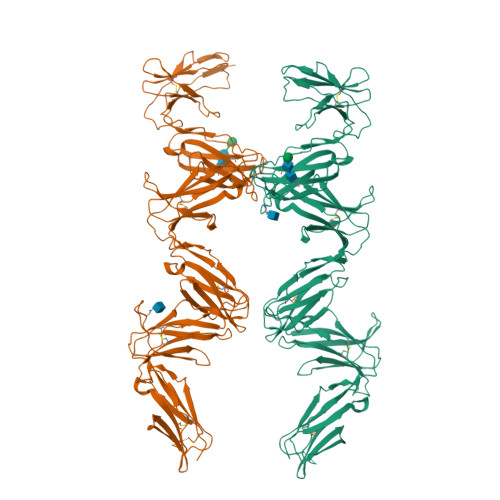
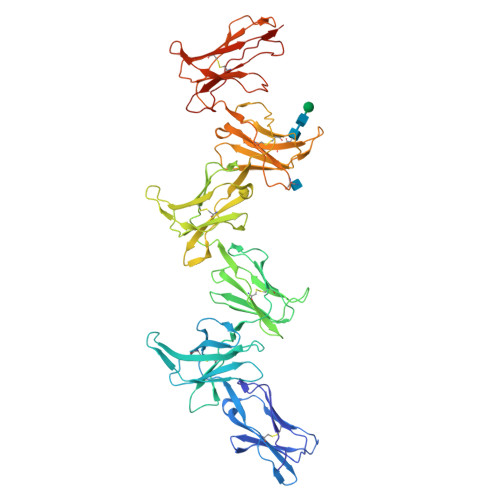
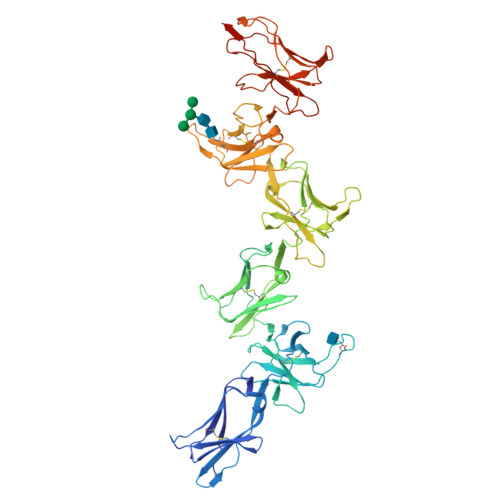
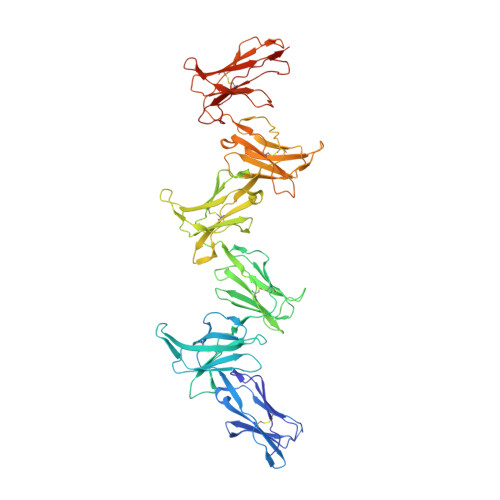
Structure and flexibility of the extracellular region of the PirB receptor.
Vlieg, H.C., Huizinga, E.G., Janssen, B.J.C.(2019) J Biological Chem 294: 4634-4643
- PubMed: 30674550
- DOI: https://doi.org/10.1074/jbc.RA118.004396
- Primary Citation of Related Structures:
6GRQ, 6GRS, 6GRT - PubMed Abstract:
Murine paired immunoglobulin receptor B (PirB) and its human ortholog leukocyte immunoglobulin-like receptor B2 (LILRB2) are widely expressed inhibitory receptors that interact with a diverse set of extracellular ligands and exert functions ranging from down-regulation of immune responses to inhibition of neuronal growth. However, structural information that could shed light on how PirB interacts with its ligands is lacking. Here, we report crystal structures of the PirB ectodomain; the first full ectodomain structure for a LILR family member, at 3.3-4.5 Å resolution. The structures reveal that PirB's six Ig-like domains are arranged at acute angles, similar to the structures of leukocyte immunoglobulin-like receptor (LILR) and killer-cell immunoglobulin-like receptor (KIR). We observe that this regular arrangement is followed throughout the ectodomain, resulting in an extended zigzag conformation. In two out of the five structures reported here, the repeating zigzag is broken by the first domain that can adopt two alternative orientations. Quantitative binding experiments revealed a 9 μm dissociation constant for PirB-myelin-associated glycoprotein (MAG) ectodomain interactions. Taken together, these structural findings and the observed PirB-MAG interactions are compatible with a model for intercellular signaling in which the PirB extracellular domains, which point away from the cell surface, enable interaction with ligands in trans .
Organizational Affiliation:
From Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, 3584 CH Utrecht, The Netherlands.