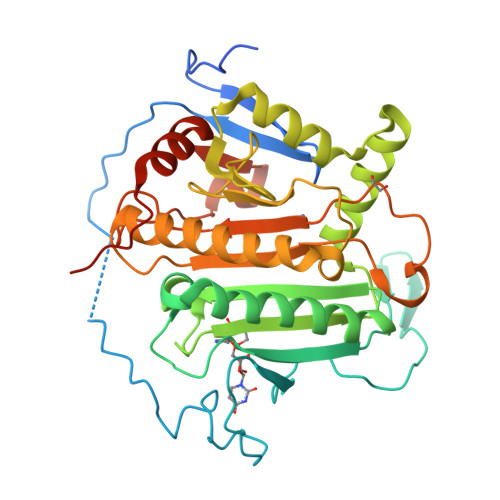
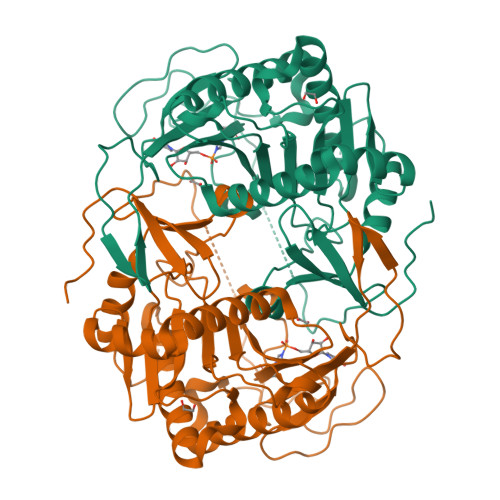
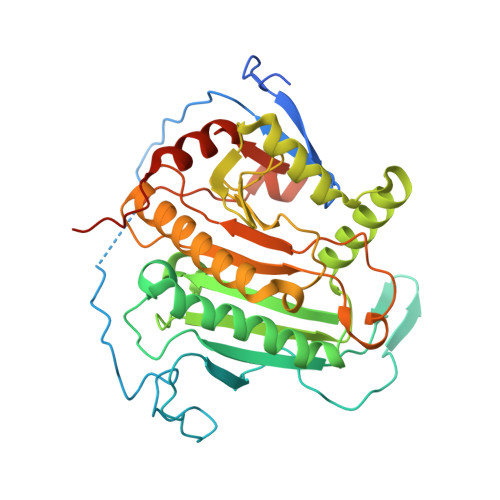
Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L
Fairhead, M., Strain-Damerell, C., Kopec, J., Bezerra, G.A., Zhang, M., Burgess-Brown, N., von Delft, F., Arrowsmith, C., Edwards, A., Bountra, C., Yue, W.W.To be published.