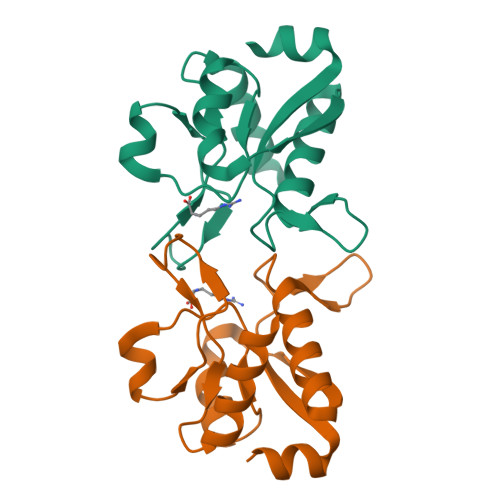
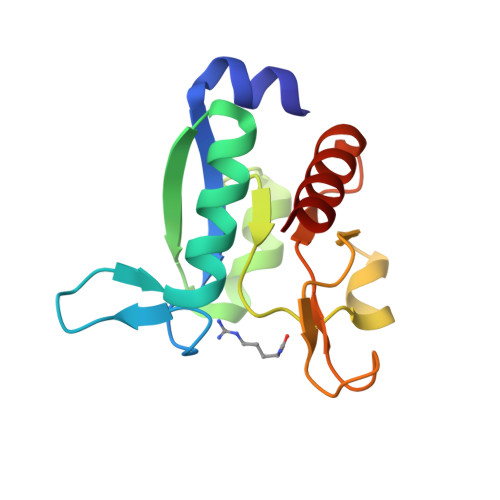
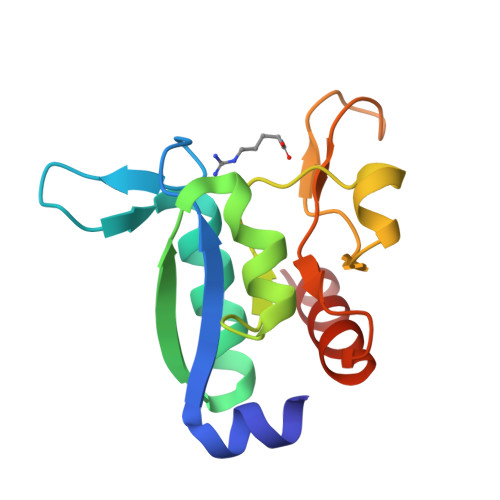
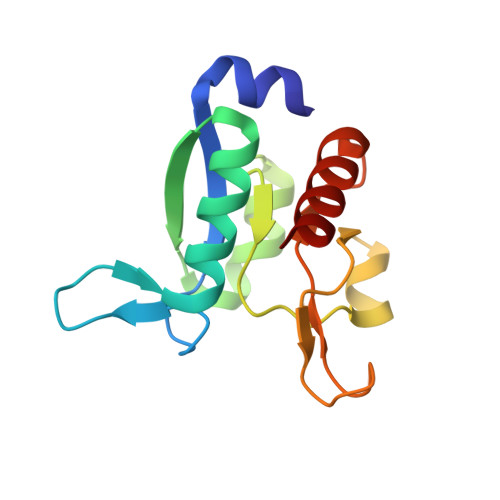
Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Smaldone, G., Balasco, N., Vigorita, M., Ruggiero, A., Cozzolino, S., Berisio, R., Del Vecchio, P., Graziano, G., Vitagliano, L.(2018) Int J Biol Macromol 119: 758-769
- PubMed: 30059738
- DOI: https://doi.org/10.1016/j.ijbiomac.2018.07.172
- Primary Citation of Related Structures:
6GPC, 6GPD, 6GPM - PubMed Abstract:
Substrate binding proteins represent a large protein family that plays fundamental roles in selective transportation of metabolites across membrane. The function of these proteins relies on the relative motions of their two domains. Insights into domain communication in this class of proteins have been here collected using Thermotoga maritima Arginine Binding Protein (TmArgBP) as model system. TmArgBP was dissected into two domains (D1 and D2) that were exhaustively characterized using a repertoire of different experimental and computational techniques. Indeed, stability, crystalline structure, ability to recognize the arginine substrate, and dynamics of the two individual domains have been here studied. Present data demonstrate that, although in the parent protein both D1 and D2 cooperate for the arginine anchoring; only D1 is intrinsically able to bind the substrate. The implications of this finding on the mechanism of arginine binding and release by TmArgBP have been discussed. Interestingly, both D1 and D2 retain the remarkable thermal/chemical stability of the parent protein. The analysis of the structural and dynamic properties of TmArgBP and of the individual domains highlights possible routes of domain communication. Finally, this study generated two interesting molecular tools, the two stable isolated domains that could be used in future investigations.
Organizational Affiliation:
IRCCS SDN, Via Emanuele Gianturco 113, 80143 Napoli, Italy. Electronic address: gsmaldone@sdn-napoli.it.