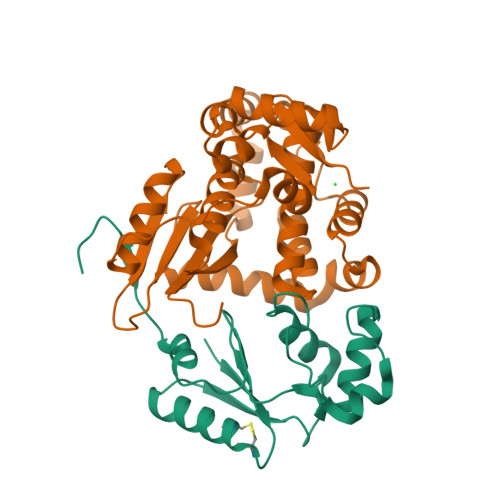
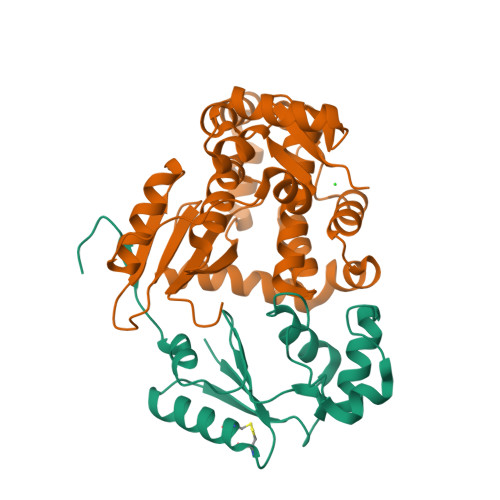
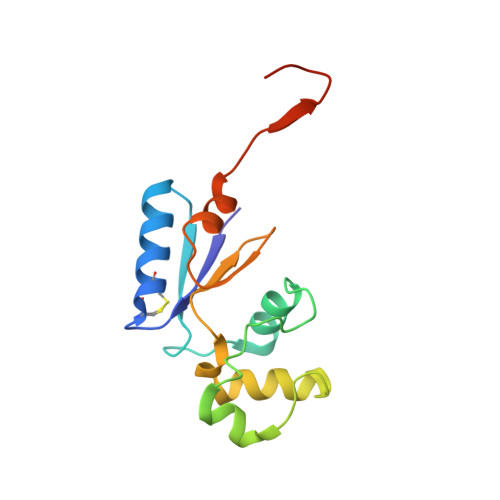
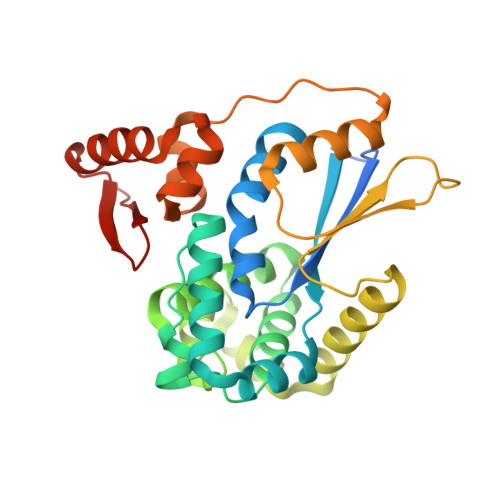
Structural Basis for YjbH Adaptor-Mediated Recognition of Transcription Factor Spx.
Awad, W., Al-Eryani, Y., Ekstrom, S., Logan, D.T., von Wachenfeldt, C.(2019) Structure 27: 923
- PubMed: 30982633
- DOI: https://doi.org/10.1016/j.str.2019.03.009
- Primary Citation of Related Structures:
6GHB, 6GHO - PubMed Abstract:
YjbH is a bacterial adaptor protein required for efficient proteolysis of the RNA polymerase-binding transcription factor Spx by the ClpXP protease. We report the structure of YjbH in complex with Spx. YjbH comprises a DsbA-like thioredoxin domain connected via a linker to a C-terminal domain reminiscent of the winged helix-turn-helix fold. The interaction between YjbH and Spx involves a large surface area. Binding to YjbH stabilizes the C-terminal ClpX recognition region of Spx. We show that mutation of critical YjbH contact residues abrogates Spx recognition. Small-angle X-ray scattering and hydrogen-deuterium exchange mass spectrometry analyses determined the existence of a stable heterodimeric complex in solution and provide evidence that binding of Spx to YjbH reduces the overall conformational flexibility of Spx. Our findings provide insights into the molecular basis for Spx recognition and suggest a model for how YjbH stabilizes Spx and displays the C terminus of Spx for engagement by ClpXP.
Organizational Affiliation:
The Microbiology Group, Department of Biology, Lund University, Sölvegatan 35, 223 62 Lund, Sweden; Department of Biophysics, Faculty of Science, Cairo University, 12316 Cairo, Egypt.