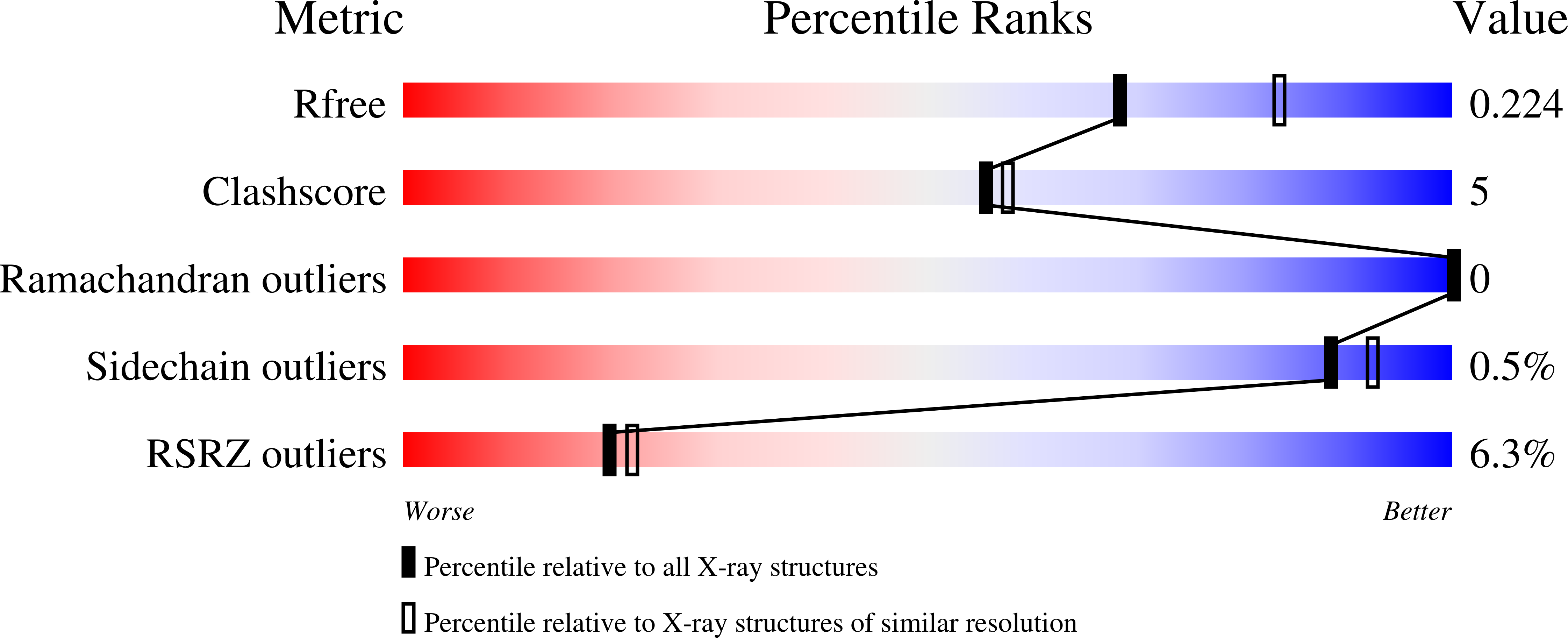
Tripeptide binding in a proton-dependent oligopeptide transporter.
Martinez Molledo, M., Quistgaard, E.M., Low, C.(2018) FEBS Lett 592: 3239-3247
- PubMed: 30194725
- DOI: https://doi.org/10.1002/1873-3468.13246
- Primary Citation of Related Structures:
6GHJ - PubMed Abstract:
Proton-dependent oligopeptide transporters (POTs) are important for the uptake of di-/tripeptides in many organisms and for drug transport in humans. The binding mode of dipeptides has been well described. However, it is still debated how tripeptides are recognized. Here, we show that tripeptides of the sequence Phe-Ala-Xxx bind with similar affinities as dipeptides to the POT transporter from Streptococcus thermophilus (PepT S t ). We furthermore determined a 2.3-Å structure of PepT S t in complex with Phe-Ala-Gln. The phenylalanine and alanine residues of the peptide adopt the same positions as previously observed for the Phe-Ala dipeptide, while the glutamine side chain extends into a hitherto uncharacterized pocket. This pocket is adaptable in size and can likely accommodate a wide variety of peptide side chains.
Organizational Affiliation:
Centre for Structural Systems Biology (CSSB), DESY and European Molecular Biology Laboratory Hamburg, Hamburg, Germany.