Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Kuhaudomlarp, S., Walpole, S., Stevenson, C.E.M., Nepogodiev, S.A., Lawson, D.M., Angulo, J., Field, R.A.(2019) Chembiochem 20: 181-192
- PubMed: 29856496
- DOI: https://doi.org/10.1002/cbic.201800260
- Primary Citation of Related Structures:
6GGY, 6GH2, 6GH3 - PubMed Abstract:
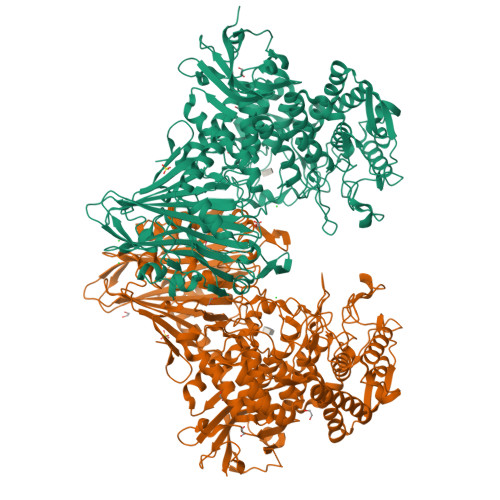
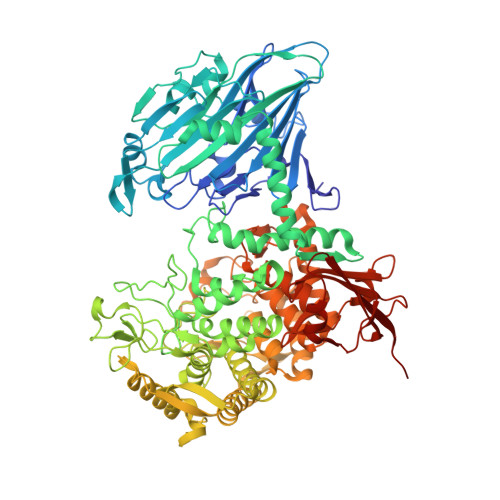
Glycoside phosphorylases (GPs) carry out a reversible phosphorolysis of carbohydrates into oligosaccharide acceptors and the corresponding sugar 1-phosphates. The reversibility of the reaction enables the use of GPs as biocatalysts for carbohydrate synthesis. Glycosyl hydrolase family 94 (GH94), which only comprises GPs, is one of the most studied GP families that have been used as biocatalysts for carbohydrate synthesis, in academic research and in industrial production. Understanding the mechanism of GH94 enzymes is a crucial step towards enzyme engineering to improve and expand the applications of these enzymes in synthesis. In this work with a GH94 laminaribiose phosphorylase from Paenibacillus sp. YM-1 (PsLBP), we have demonstrated an enzymatic synthesis of disaccharide 1 (β-d-mannopyranosyl-(1→3)-d-glucopyranose) by using a natural acceptor glucose and noncognate donor substrate α-mannose 1-phosphate (Man1P). To investigate how the enzyme recognises different sugar 1-phosphates, the X-ray crystal structures of PsLBP in complex with Glc1P and Man1P have been solved, providing the first molecular detail of the recognition of a noncognate donor substrate by GPs, which revealed the importance of hydrogen bonding between the active site residues and hydroxy groups at C2, C4, and C6 of sugar 1-phosphates. Furthermore, we used saturation transfer difference NMR spectroscopy to support crystallographic studies on the sugar 1-phosphates, as well as to provide further insights into the PsLBP recognition of the acceptors and disaccharide products.
Organizational Affiliation:
Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK.