Crystal structure of an ancient sequence-reconstructed Elongation Factor Tu (node 262)
Majumdar, S., Tarafder, A.D., Ge, X., Kacar, B., Sanyal, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Elongation Factor Tu | 400 | synthetic construct | Mutation(s): 0 | 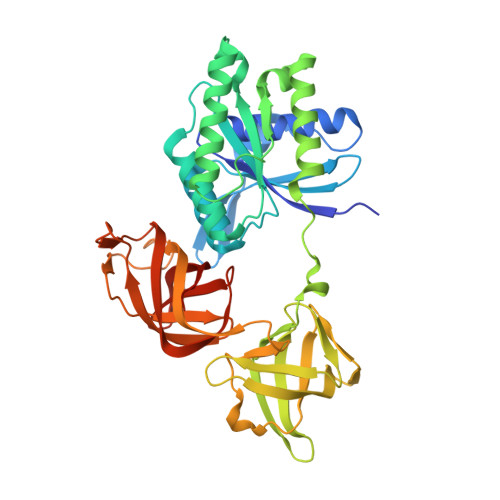 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GDP Query on GDP | B [auth A] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| MG Query on MG | C [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.944 | α = 90 |
| b = 73.691 | β = 90 |
| c = 113.756 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Auto-Rickshaw | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swedish Research Council | Sweden | 2014-4423 |
| Swedish Research Council | Sweden | 2016-06264 |
| Wenner-Gren Foundation | Sweden | UPD 2017-0238 |
| Knut and Alice Wallenberg Foundation | Sweden | KAW 2017.0055 |