Structure-based optimization of bisphosphonate nucleoside inhibitors of human 5'(3')-deoxyribonucleotidases
Pachl, P., Simak, O., Budesinsky, M., Brynda, J., Rosenberg, I., Rezacova, P.(2018) European J Org Chem
Experimental Data Snapshot
Starting Model: experimental
View more details
(2018) European J Org Chem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'(3')-deoxyribonucleotidase, mitochondrial | 202 | Homo sapiens | Mutation(s): 0 Gene Names: NT5M, DNT2 EC: 3.1.3 | 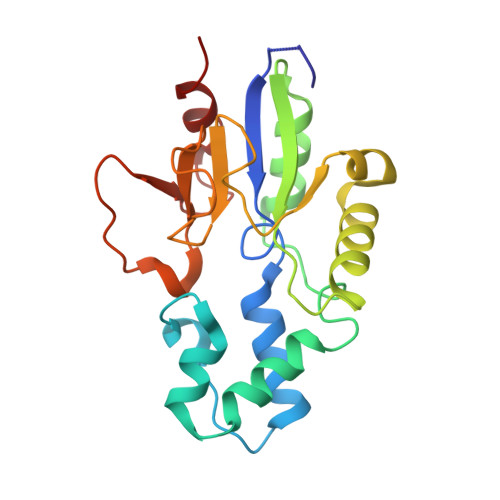 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NPB1 (Homo sapiens) Explore Q9NPB1 Go to UniProtKB: Q9NPB1 | |||||
PHAROS: Q9NPB1 GTEx: ENSG00000205309 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NPB1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| O84 Query on O84 | F [auth A] | [(2~{R},4~{a}~{R},6~{R},7~{a}~{R})-6-[2,4-bis(oxidanylidene)-5-[(~{E})-3-phosphonoprop-1-enyl]pyrimidin-1-yl]-2-phenyl-4~{a},6,7,7~{a}-tetrahydro-4~{H}-furo[3,2-d][1,3]dioxin-2-yl]phosphonic acid C19 H22 N2 O11 P2 TZXVMSWLUKAZDT-PDBOZARJSA-N |  | ||
| GOL Query on GOL | C [auth A], D [auth A], E [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| BME Query on BME | B [auth A] | BETA-MERCAPTOETHANOL C2 H6 O S DGVVWUTYPXICAM-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | G [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| K Query on K | I [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | H [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.858 | α = 90 |
| b = 73.858 | β = 90 |
| c = 106.121 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Grant Agency of the Czech Republic | Czech Republic | 203-09-0820 |
| Grant Agency of the Czech Republic | Czech Republic | 15-05677S |
| Czech Science Foundation | Czech Republic | 17-12703S |