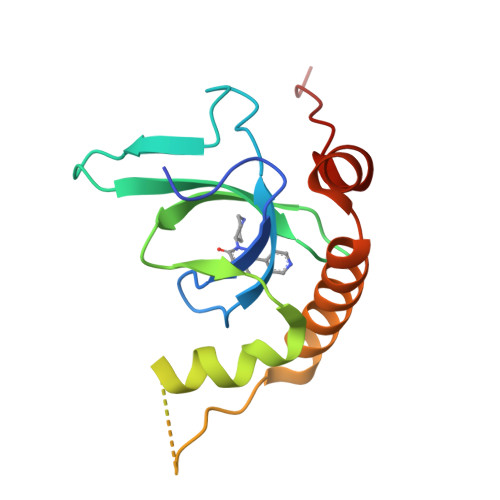
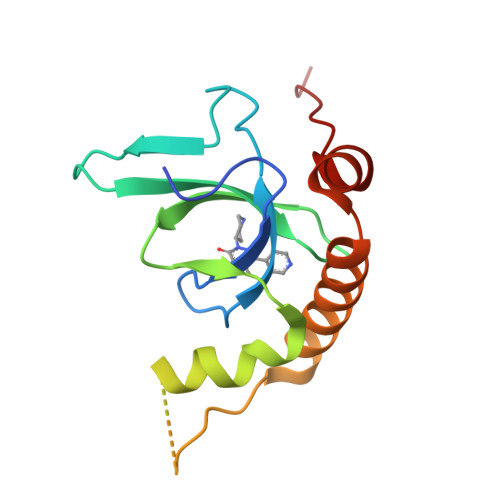
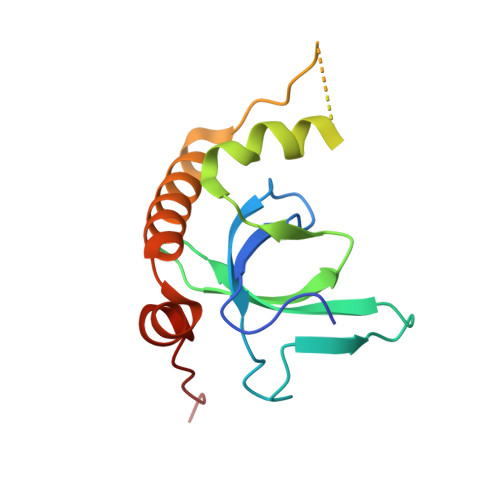
Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Bottcher, J., Dilworth, D., Reiser, U., Neumuller, R.A., Schleicher, M., Petronczki, M., Zeeb, M., Mischerikow, N., Allali-Hassani, A., Szewczyk, M.M., Li, F., Kennedy, S., Vedadi, M., Barsyte-Lovejoy, D., Brown, P.J., Huber, K.V.M., Rogers, C.M., Wells, C.I., Fedorov, O., Rumpel, K., Zoephel, A., Mayer, M., Wunberg, T., Bose, D., Zahn, S., Arnhof, H., Berger, H., Reiser, C., Hormann, A., Krammer, T., Corcokovic, M., Sharps, B., Winkler, S., Haring, D., Cockcroft, X.L., Fuchs, J.E., Mullauer, B., Weiss-Puxbaum, A., Gerstberger, T., Boehmelt, G., Vakoc, C.R., Arrowsmith, C.H., Pearson, M., McConnell, D.B.(2019) Nat Chem Biol 15: 822-829
- PubMed: 31285596
- DOI: https://doi.org/10.1038/s41589-019-0310-x
- Primary Citation of Related Structures:
6G24, 6G25, 6G27, 6G29, 6G2B, 6G2C, 6G2E, 6G2F, 6G2O, 6G3P, 6G3T - PubMed Abstract:
Here, we report the fragment-based discovery of BI-9321, a potent, selective and cellular active antagonist of the NSD3-PWWP1 domain. The human NSD3 protein is encoded by the WHSC1L1 gene located in the 8p11-p12 amplicon, frequently amplified in breast and squamous lung cancer. Recently, it was demonstrated that the PWWP1 domain of NSD3 is required for the viability of acute myeloid leukemia cells. To further elucidate the relevance of NSD3 in cancer biology, we developed a chemical probe, BI-9321, targeting the methyl-lysine binding site of the PWWP1 domain with sub-micromolar in vitro activity and cellular target engagement at 1 µM. As a single agent, BI-9321 downregulates Myc messenger RNA expression and reduces proliferation in MOLM-13 cells. This first-in-class chemical probe BI-9321, together with the negative control BI-9466, will greatly facilitate the elucidation of the underexplored biological function of PWWP domains.
Organizational Affiliation:
Boehringer Ingelheim RCV GmbH & Co KG, Vienna, Austria. jark.boettcher@boehringer-ingelheim.com.