Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
Ahangar, M.S., Furze, C.M., Guy, C.S., Cooper, C., Maskew, K.S., Graham, B., Cameron, A.D., Fullam, E.(2018) J Biol Chem 293: 9770-9783
- PubMed: 29728457
- DOI: https://doi.org/10.1074/jbc.RA118.002597
- Primary Citation of Related Structures:
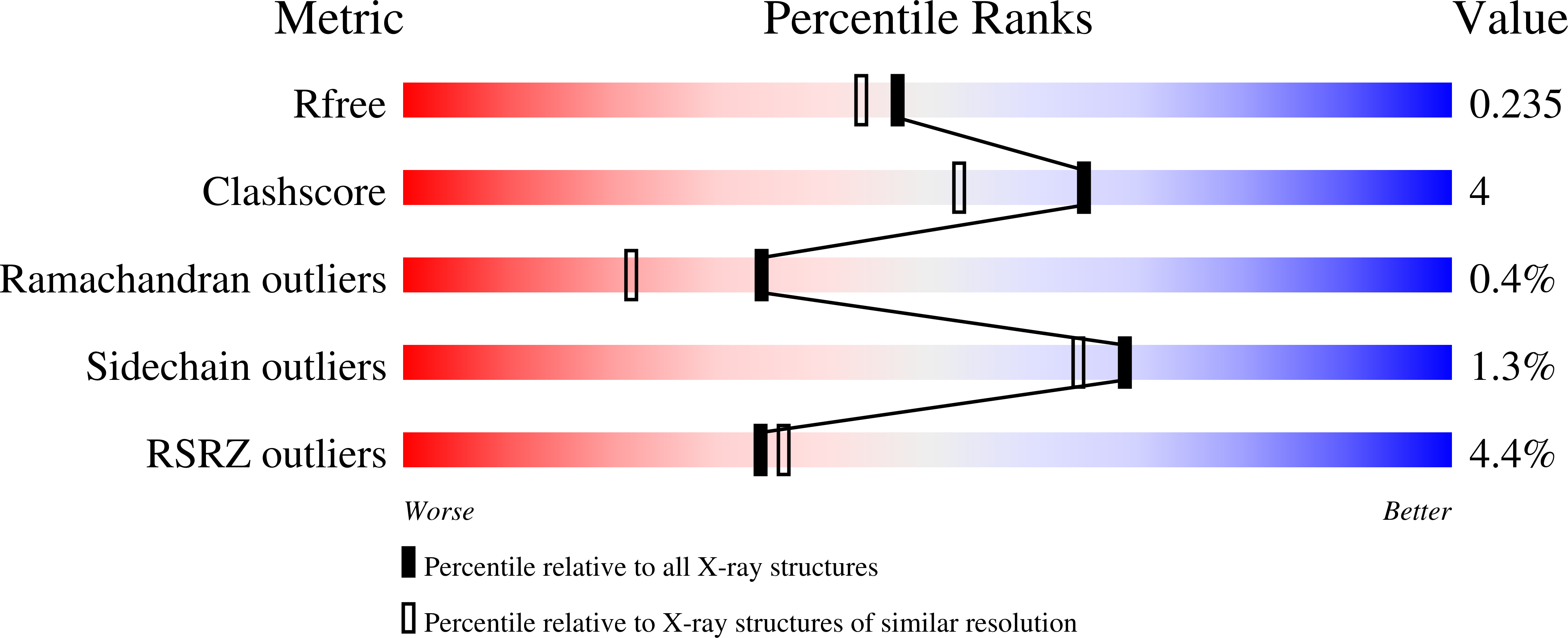
6FV3, 6FV4 - PubMed Abstract:
The Mycobacterium tuberculosis ( Mtb ) pathogen encodes a GlcNAc-6-phosphate deacetylase enzyme, NagA (Rv3332), that belongs to the amidohydrolase superfamily. NagA enzymes catalyze the deacetylation of GlcNAc-6-phosphate (GlcNAc6P) to glucosamine-6-phosphate (GlcN6P). NagA is a potential antitubercular drug target because it represents the key enzymatic step in the generation of essential amino-sugar precursors required for Mtb cell wall biosynthesis and also influences recycling of cell wall peptidoglycan fragments. Here, we report the structural and functional characterization of NagA from Mycobacterium smegmatis (MSNagA) and Mycobacterium marinum (MMNagA), close relatives of Mtb Using a combination of X-ray crystallography, site-directed mutagenesis, and biochemical and biophysical assays, we show that these mycobacterial NagA enzymes are selective for GlcNAc6P. Site-directed mutagenesis studies revealed crucial roles of conserved residues in the active site that underpin stereoselective recognition, binding, and catalysis of substrates. Moreover, we report the crystal structure of MSNagA in both ligand-free form and in complex with the GlcNAc6P substrate at 2.6 and 2.0 Å resolutions, respectively. The GlcNAc6P complex structure disclosed the precise mode of GlcNAc6P binding and the structural framework of the active site, including two divalent metals located in the α/β binuclear site. Furthermore, we observed a cysteine residue located on a flexible loop region that occludes the active site. This cysteine is unique to mycobacteria and may represent a unique subsite for targeting mycobacterial NagA enzymes. Our results provide critical insights into the structural and mechanistic properties of mycobacterial NagA enzymes having an essential role in amino-sugar and nucleotide metabolism in mycobacteria.
Organizational Affiliation:
From the School of Life Sciences and.