NMR Structure Determinations of Small Proteins Using only One Fractionally 20% 13 C- and Uniformly 100% 15 N-Labeled Sample.
Heikkinen, H.A., Backlund, S.M., Iwai, H.(2021) Molecules 26
- PubMed: 33535444
- DOI: https://doi.org/10.3390/molecules26030747
- Primary Citation of Related Structures:
6FFQ, 6FFU, 6SOW - PubMed Abstract:
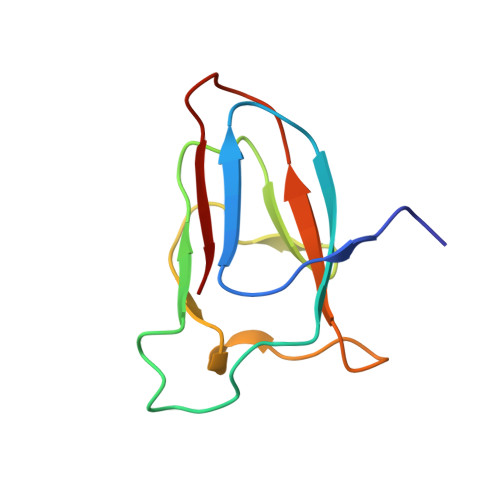
Uniformly 13 C- and 15 N-labeled samples ensure fast and reliable nuclear magnetic resonance (NMR) assignments of proteins and are commonly used for structure elucidation by NMR. However, the preparation of uniformly labeled samples is a labor-intensive and expensive step. Reducing the portion of 13 C-labeled glucose by a factor of five using a fractional 20% 13 C- and 100% 15 N-labeling scheme could lower the total chemical costs, yet retaining sufficient structural information of uniformly [ 13 C, 15 N]-labeled sample as a result of the improved sensitivity of NMR instruments. Moreover, fractional 13 C-labeling can facilitate reliable resonance assignments of sidechains because of the biosynthetic pathways of each amino-acid. Preparation of only one [20% 13 C, 100% 15 N]-labeled sample for small proteins (<15 kDa) could also eliminate redundant sample preparations of 100% 15 N-labeled and uniformly 100% [ 13 C, 15 N]-labeled samples of proteins. We determined the NMR structures of a small alpha-helical protein, the C domain of IgG-binding protein A from Staphylococcus aureus (SpaC), and a small beta-sheet protein, CBM64 module using [20% 13 C, 100% 15 N]-labeled sample and compared with the crystal structures and the NMR structures derived from the 100% [ 13 C, 15 N]-labeled sample. Our results suggest that one [20% 13 C, 100% 15 N]-labeled sample of small proteins could be routinely used as an alternative to conventional 100% [ 13 C, 15 N]-labeling for backbone resonance assignments, NMR structure determination, 15 N-relaxation analysis, and ligand-protein interaction.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki. P.O. Box 65, FIN-00014 Helsinki, Finland.