Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
Yang, B., Vasbinder, M.M., Hird, A.W., Su, Q., Wang, H., Yu, Y., Toader, D., Lyne, P.D., Read, J.A., Breed, J., Ioannidis, S., Deng, C., Grondine, M., DeGrace, N., Whitston, D., Brassil, P., Janetka, J.W.(2018) J Med Chem 61: 1061-1073
- PubMed: 29301085
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01490
- Primary Citation of Related Structures:
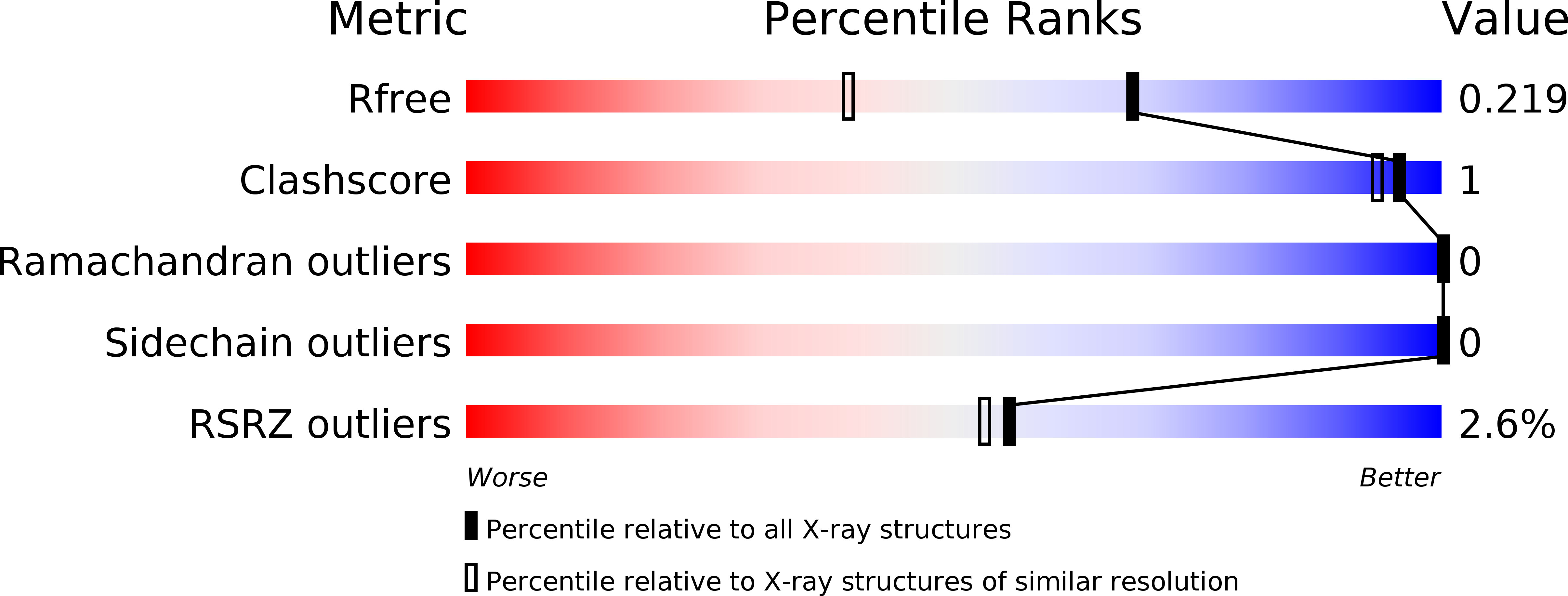
6FC8, 6FCF, 6FCK - PubMed Abstract:
Checkpoint kinase 1 (CHK1) inhibitors are potential cancer therapeutics that can be utilized for enhancing the efficacy of DNA damaging agents. Multiple small molecule CHK1 inhibitors from different chemical scaffolds have been developed and evaluated in clinical trials in combination with chemotherapeutics and radiation treatment. Scaffold morphing of thiophene carboxamide ureas (TCUs), such as AZD7762 (1) and a related series of triazoloquinolines (TZQs), led to the identification of fused-ring bicyclic CHK1 inhibitors, 7-carboxamide thienopyridines (7-CTPs), and 7-carboxamide indoles. X-ray crystal structures reveal a key intramolecular noncovalent sulfur-oxygen interaction in aligning the hinge-binding carboxamide group to the thienopyridine core in a coplanar fashion. An intramolecular hydrogen bond to an indole NH was also effective in locking the carboxamide in the preferred bound conformation to CHK1. Optimization on the 7-CTP series resulted in the identification of lead compound 44, which displayed respectable drug-like properties and good in vitro and in vivo potency.
Organizational Affiliation:
Oncology Chemistry, IMED Biotech Unit, AstraZeneca , 35 Gatehouse Drive, Waltham, Massachusetts 02451, United States.