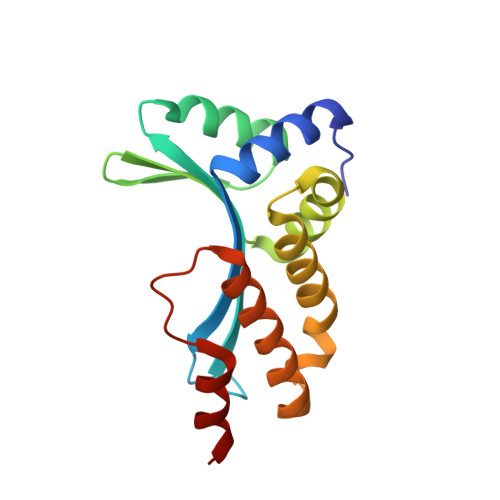
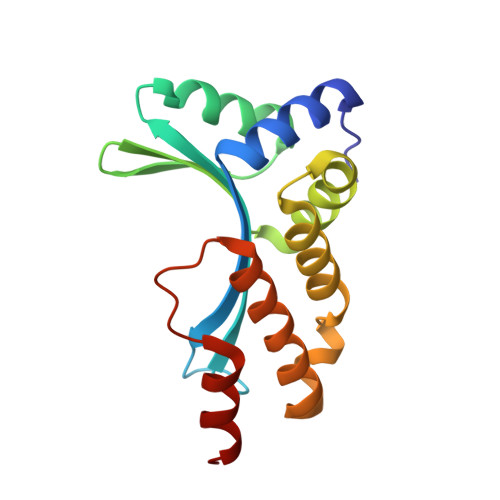
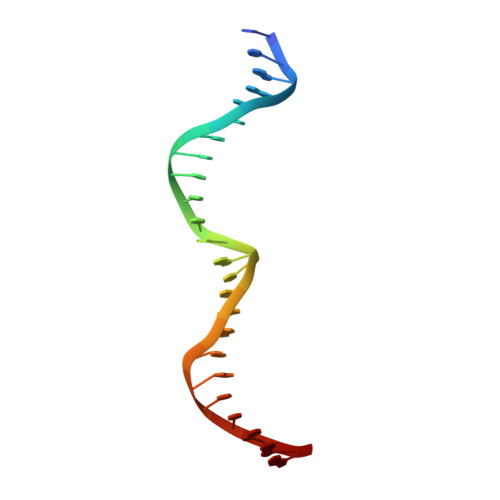
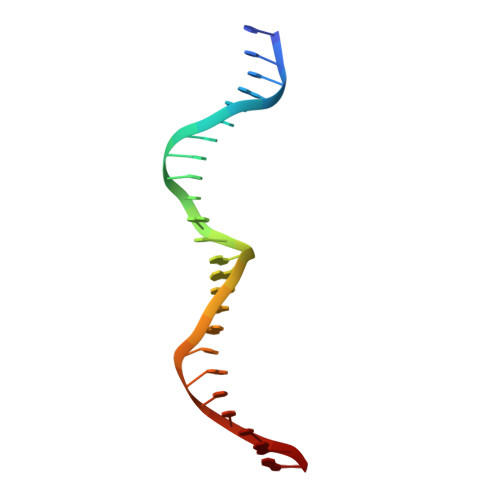
Understanding the indirect DNA read-out specificity of I-CreI Meganuclease.
Prieto, J., Redondo, P., Lopez-Mendez, B., D'Abramo, M., Merino, N., Blanco, F.J., Duchateau, P., Montoya, G., Molina, R.(2018) Sci Rep 8: 10286-10286
- PubMed: 29980759
- DOI: https://doi.org/10.1038/s41598-018-28599-0
- Primary Citation of Related Structures:
6FB0, 6FB1, 6FB2, 6FB5, 6FB6, 6FB7, 6FB8, 6FB9 - PubMed Abstract:
The high DNA specificity of homing endonucleases makes them a powerful protein scaffold to engineer enzymes for genome manipulation. Understanding their molecular recognition of DNA is an important prerequisite to generate engineered enzymes able to cleave DNA in specific desired genome sites. Protein-DNA recognition studies have been mostly focused on specific direct contacts between amino acid side chains and bases to redesign the binding interface. However, the important role of indirect readout in the central region of the target DNA of the homing endonuclease I-CreI suggested that indirect readout may play a key role in the redesign of protein-DNA interactions. The sequences of the I-CreI central substrate region, 2NN, along with the adjacent 5NNN, are key for substrate cleavage. Here, we analyse the mechanism of target discrimination at the 5NNN region by the I-CreI protein, revealing its critical role in the location and occupancy of the catalytic metal ions, which is crucial for cleavage. Our data highlight the importance of indirect readout for target DNA cleavage, thus aiding I-CreI engineering when targeting new DNA sequences.
Organizational Affiliation:
Structural Biology and Biocomputing Programme, Spanish National Cancer Research Centre (CNIO), Macromolecular Crystallography Group, c/Melchor Fdez. Almagro 3, 28029, Madrid, Spain.