Kinetic Resolution of sec-Thiols by Enantioselective Oxidation with Rationally Engineered 5-(Hydroxymethyl)furfural Oxidase.
Pickl, M., Swoboda, A., Romero, E., Winkler, C.K., Binda, C., Mattevi, A., Faber, K., Fraaije, M.W.(2018) Angew Chem Int Ed Engl 57: 2864-2868
- PubMed: 29384246
- DOI: https://doi.org/10.1002/anie.201713189
- Primary Citation of Related Structures:
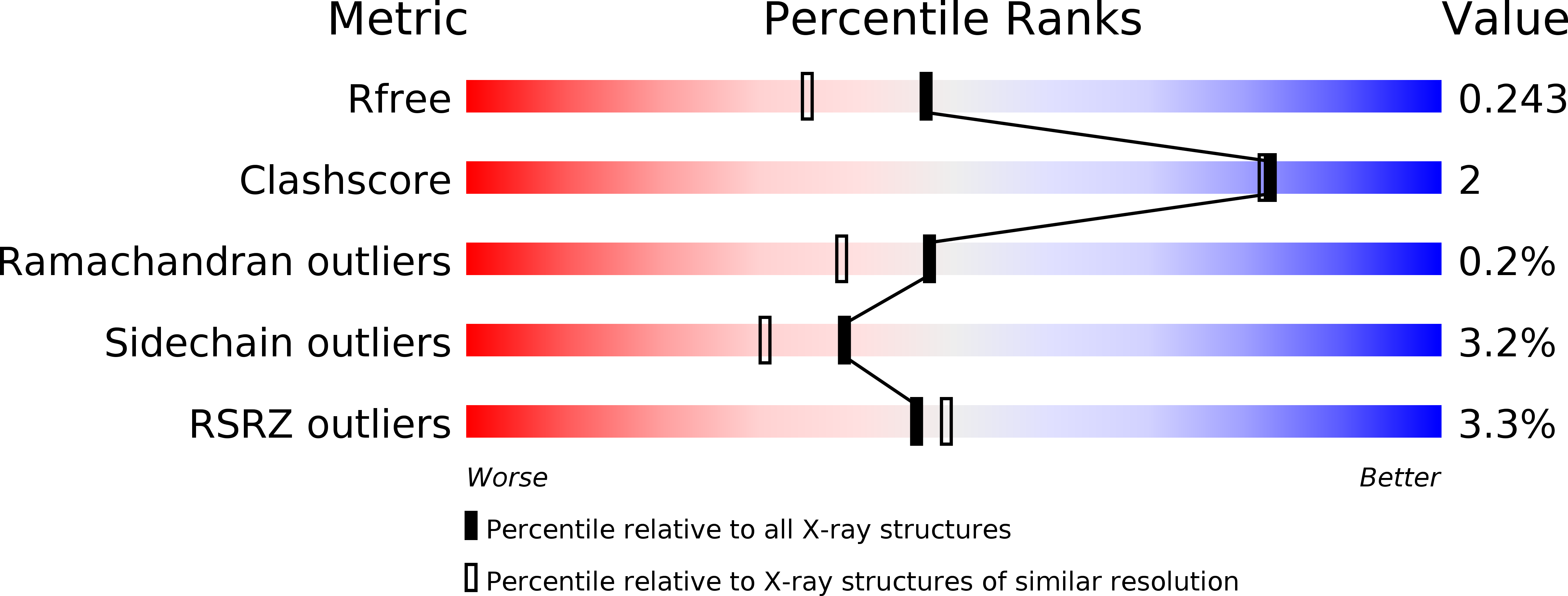
6F97 - PubMed Abstract:
Various flavoprotein oxidases were recently shown to oxidize primary thiols. Herein, this reactivity is extended to sec-thiols by using structure-guided engineering of 5-(hydroxymethyl)furfural oxidase (HMFO). The variants obtained were employed for the oxidative kinetic resolution of racemic sec-thiols, thus yielding the corresponding thioketones and nonreacted R-configured thiols with excellent enantioselectivities (E≥200). The engineering strategy applied went beyond the classic approach of replacing bulky amino acid residues with smaller ones, as the active site was additionally enlarged by a newly introduced Thr residue. This residue established a hydrogen-bonding interaction with the substrates, as verified in the crystal structure of the variant. These strategies unlocked HMFO variants for the enantioselective oxidation of a range of sec-thiols.
Organizational Affiliation:
Department of Chemistry, University of Graz, Heinrichstrasse 28, 8010, Graz, Austria.